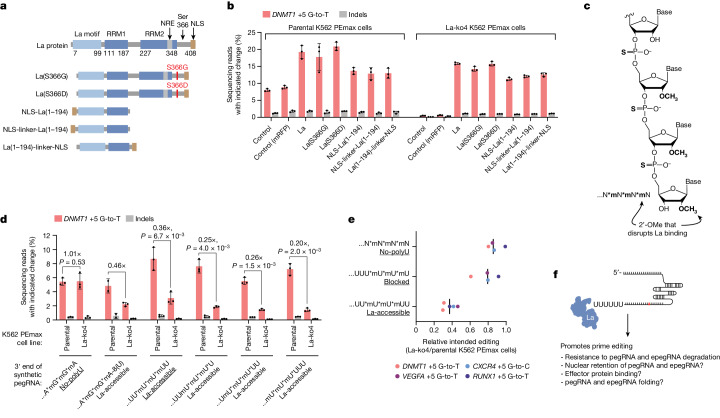
Fig. 3. La functionally interacts with the 3′ ends of polyuridylated pegRNAs.
a, Domain architectures of La and mutants. NRE, nuclear retention element Linker, SGGS. b, Percentages of prime editing outcomes with or without ectopic expression of La or mutants depicted in a. Expression plasmids were delivered to indicated cells alongside the plasmid encoding the DNMT1 +5 G-to-T pegRNA. c, Schematic of RNA with chemical modifications (bold); specifically, phosphorothioate bonds (asterisks in sequence representation) and 2′-OMe modifications (‘m’ in sequence representation). d, Percentages of prime editing outcomes produced using 100 pmole of synthetic pegRNAs with indicated 3′ end configurations. e, Fold changes in average intended prime editing at four genomic loci in La-ko4 cells relative to parental K562 PEmax cells produced using 100 pmole of synthetic pegRNA with the indicated 3′ end configurations. Editing percentages provided in Extended Data Fig. 5e. f, Model of La interaction with pegRNAs. The PE2 approach was used in b,d,e. Underlining in d,e indicates particular 3′ end configuration patterns. Data and error bars in b and d indicate the mean ± s.d. (n = 2–3 independent biological replicates). Vertical bars in e indicate medians (4 edits) of ratios of means (n = 3 independent biological replicates for each edit). P values in d are from one-tailed unpaired Student’s t-test. Image of pegRNA in f adapted from ref. 40, Elsevier.