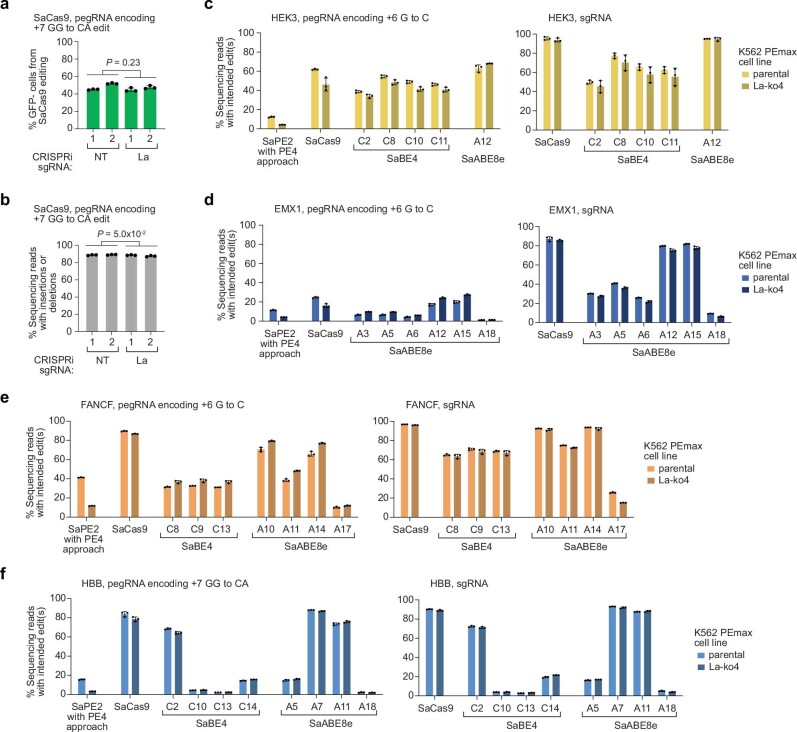
Extended Data Fig. 4. La has a stronger impact on prime editing than other editing modalities.
a, Percentages of GFP- cells within indicated cell populations arising from SaCas9-induced DSBs at a stably integrated MCS reporter in K562 CRISPRi cells. CRISPRi sgRNAs were delivered by lentiviral transduction. Editing components (SaCas9, +7 GG to CA pegRNA) were delivered by plasmid transfection. Representative flow cytometry data from each condition and unedited controls also presented in Fig. 2f. b, Quantification of SaCas9-induced indels at stably integrated MCS reporter described in a. c-f, Percentages of intended editing achieved in K562 PEmax parental and La-ko4 cells using SaPE2 with the PE4 approach, SaCas9, SaBE4, and SaABE8e across four genomic loci, HEK3 (c), EMX1 (d), FANCF (e) and HBB (f). The same pegRNA or sgRNA expression plasmid was used for all editing systems at each target, with select combinations excluded (SaPE2 with PE4 approach with any sgRNA and SaBE4 at EMX1). Relative editing for each intended outcome presented in Fig. 2g and h. Data and error bars in a-f indicate mean ± s.d. (n = 3 independent biological replicates). P-values in a and b are from two-tailed unpaired Student’s t-test.