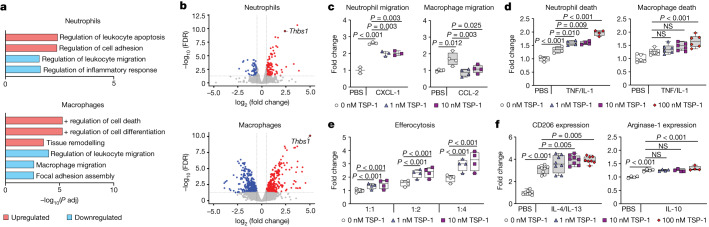
Fig. 3. CGRP upregulates TSP-1 in neutrophils and macrophages to mediate its activity.
a,b, RNA-seq analysis of CGRP-treated neutrophils and macrophages (1 nM or saline for 4 h). Differential gene expression was performed with limma-voom (false discovery rate (FDR) < 0.05). Fold change values returned by limma were used for pathway analysis with FDR < 0.05 to correct for multiple comparisons. a, GO enrichment analysis of significantly upregulated (red) and downregulated (blue) genes in CGRP-treated and saline-treated groups. b, Volcano plots showing DEGs with fold change > |1.5| between CGRP-treated and saline-treated groups. Significantly upregulated (red) and downregulated (blue) genes in CGRP-treated neutrophils and macrophages are shown (n = 3). c–f, Neutrophils and macrophages were treated with saline (PBS, 0 nM TSP-1) or TSP-1 (1, 10 or 100 nM). Results are expressed as fold change over the PBS (0 nM TSP-1) control group. Boxes show median (centre line) and interquartile range (edges), whiskers show the range of values. Dots represent independent experiments. c, Transwell migration towards a chemoattractant (CXCL1 or CCL2) after TSP-1 treatment (n = 3 for neutrophils, n = 4 for macrophages). d, Cell death in response to TSP-1 with or without TNF plus IL-1 (n = 4). e, Macrophage efferocytosis of neutrophils after TSP-1 treatment (n = 4). f, Macrophage M2-like polarization determined via CD206 and arginase-1 expression in response to TSP-1 and IL-4/IL-13 or IL-10 treatments (CD206, n = 8; arginase-1, n = 4). c–f, One-way ANOVA with Tukey post hoc test for pairwise comparisons. P values are indicated.