FIGURE 7.
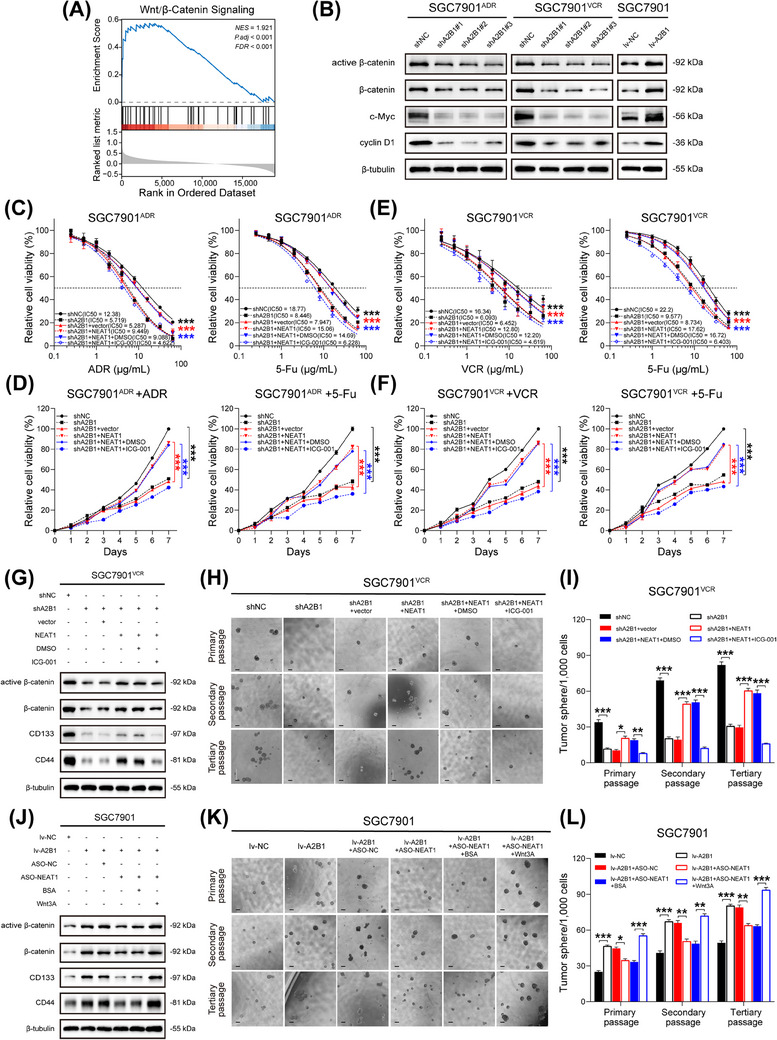
hnRNPA2B1 and NEAT1 enhanced stemness and promoted chemoresistance via Wnt/β‐catenin pathway in GC. (A) GSEA analysis between hnRNPA2B1high NEAT1high group (n = 105) and hnRNPA2B1low NEAT1low group (n = 104). (B) Immunoblots of active β‐catenin, β‐catenin, c‐Myc, and cyclin D1 in hnRNPA2B1‐silenced SGC7901ADR cells, hnRNPA2B1‐silenced SGC7901VCR cells and hnRNPA2B1‐overexpressed SGC7901 cells cell lines respectively. (C‐D) IC50 values and growth curves of SGC7901ADR cells after the indicated treatment (ICG‐001, 25 µmol/L for 24 h). (E‐F) IC50 values and growth curves of SGC7901VCR cells after the indicated treatment (ICG‐001, 25 µmol/L for 24 h). (G) Immunoblots of active β‐catenin, β‐catenin, CD133 and CD44 in SGC7901VCR cells after the indicated treatment (ICG‐001, 25 µmol/L for 24 h). (H‐I) Representative images (H) and quantification (I) of the sphere‐forming abilities of SGC7901VCR cells after the indicated treatment. Scale bar, 200 µm. (J) Immunoblots of active β‐catenin, β‐catenin, CD133 and CD44 in SGC7901 cells after the indicated treatment (Wnt3A, 100 ng/mL for 24 h). (K‐L) Representative images (K) and quantification (L) of the sphere‐forming abilities of SGC7901 cells after the indicated treatment. Scale bar, 200 µm. The P value was calculated by two‐way ANOVA test (C‐F), and paired t test (I and L). *P < 0.05, ** P < 0.01, *** P < 0.001. Abbreviations: CD133, cluster of differentiation 133; CD44, cluster of differentiation 44; FDR, false discovery rate; GSEA, gene set enrichment analysis; hnRNPA2B1, heterogeneous nuclear ribonucleoprotein A2B1; lv‐NC, empty overexpression control; lv‐A2B1, overexpression of hnRNPA2B1; NES, normalized enrichment score; P.adj, adjust P value; shNC, negative control short hairpin RNA; shA2B1, short hairpin RNA of hnRNPA2B1.
