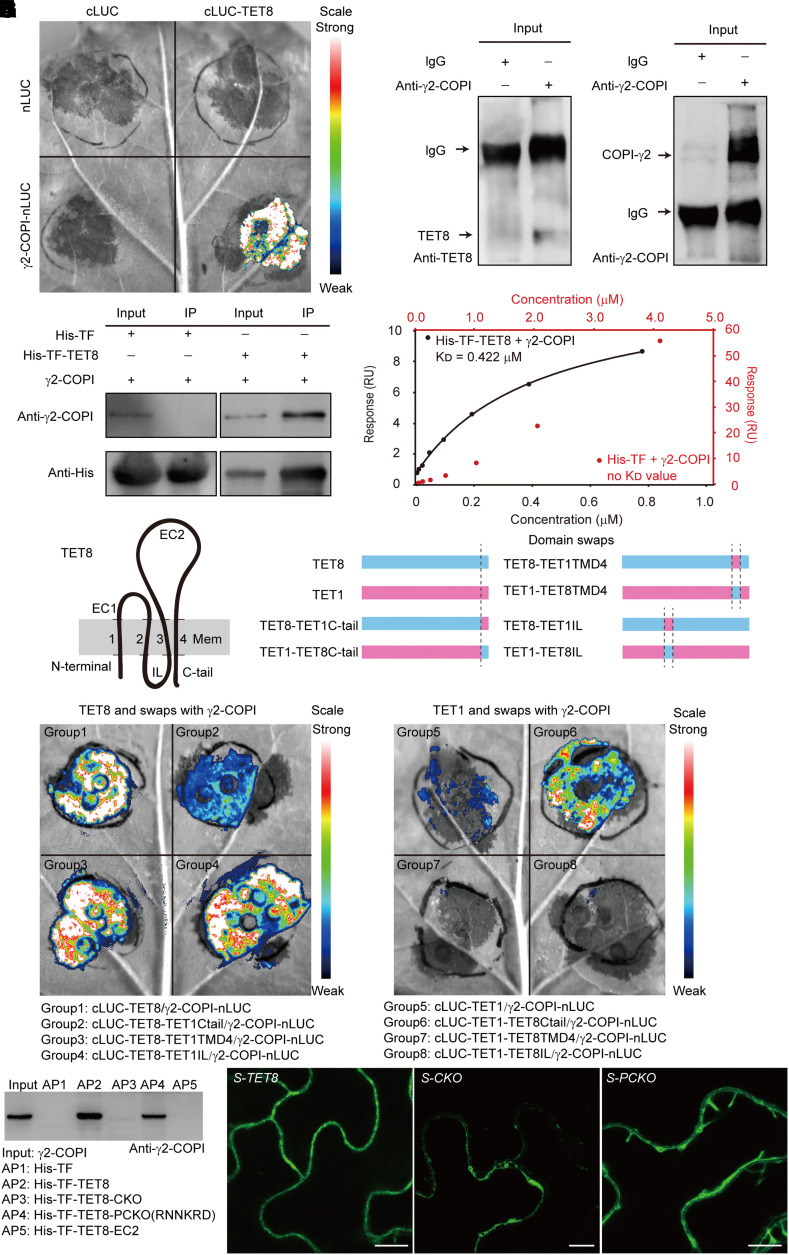
Figure 2.
The C-tail of TET8 can recognize γ2-COPI and determines proper TET8 localization. A) Bimolecular luminescence complementation (BiLC) assay showing that TET8 interacts with γ2-COPI strongly. Luminescence intensities indicate the degree of binding capacity. B) Co-IP assays showing that TET8 interacts with γ2-COPI in vivo. Beads incubated with IgG were used as negative control. C) Pull-down assays demonstrating that recombinant His-TF-TET8 binds to γ2-COPI in vitro. Beads incubated with recombinant His-TF were used as negative control. For B) and C), same amounts of total proteins or bait proteins were loaded. D) SPR assay showing that His-TF-TET8 can bind to γ2-COPI. The KD (affinity) value was calculated based on the fitting curves and represents the binding affinity. E) Topology of TET8 in the membrane. The transmembrane domains of TET8 are numbered 1 to 4. EC1, extracellular loop 1; EC2, extracellular loop 2, IL, internal loop; C-tail, C-terminal tail. F) Diagrams illustrating the domain swaps between TET1 and TET8. G, H) BiLC assays showing the interaction between recombinant TET proteins with γ2-COPI. Fluorescence intensities indicate the degree of binding capacity. I) Pull-down assays showing the direct interaction between TET8-C-tail and γ2-COPI. AP, affinity precipitate. The same amounts of proteins were loaded in each lane. J) Cellular distribution of EGFP-TET8, EGFP-TET8-CKO, and EGFP-TET8-PCKO in transgenic plants. Scale bars, 10 μm.