Fig. 3 |. In vitro optimization of C3 LNPs for co-delivery of base editing platforms.
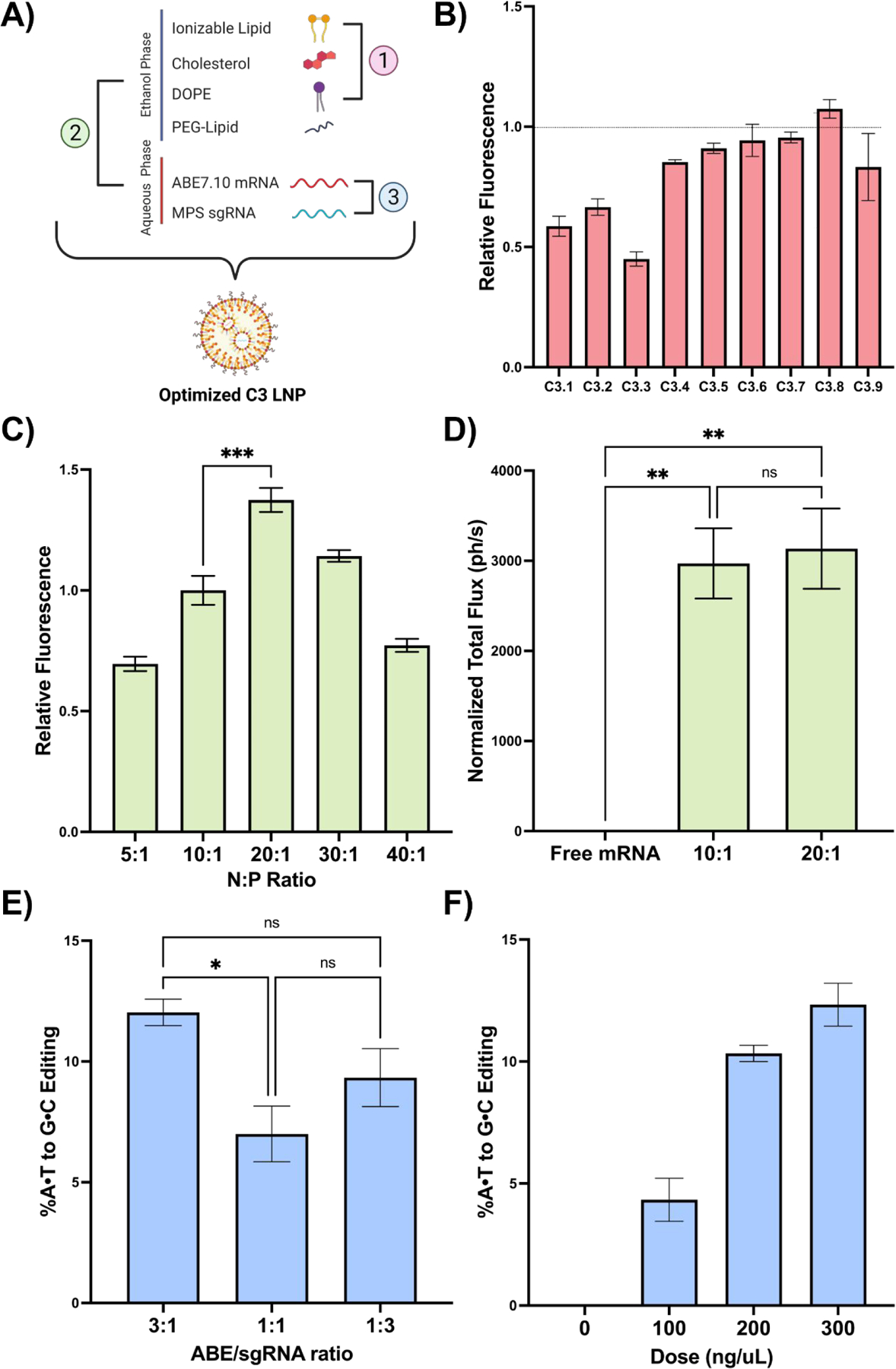
(A) Scheme demonstrating the three LNP formulation parameters evaluated: 1) excipient molar ratios, 2) N:P ratio, and 3) ABE to sgRNA mass ratio. (B) Normalized mean fluorescence intensity (MFI) in Neuro-2a cells after treatment with the C3 LNP DOE library relative to the original C3 LNP formulation (dotted line). (C) Normalized MFI in Neuro-2a cells after treatment with C3 LNPs formulated at a range of N:P ratios. (D) Quantification of luminescence signal (normalized total flux) from brains of neonates treated with C3 LNPs prepared at the top-performing N:P ratios in vitro. (E) Sequencing results at the expected site of base editing following C3 LNP-mediated base editing in primary MPS-I murine fibroblasts at three ABE/sgRNA mass ratios. (F) Sequencing results in primary MPS-I murine neurons treated with C3.MPS LNPs at a range of doses. * p < 0.05, ** p < 0.01, *** p < 0.001 by one-way ANOVA with post-hoc Dunnett’s test compared positive controls described in the text. Outliers were detected using Grubbs’ test and removed from analysis; minimum n = 3 replicates per treatment group; error bars represent SEM.
