Figure 1.
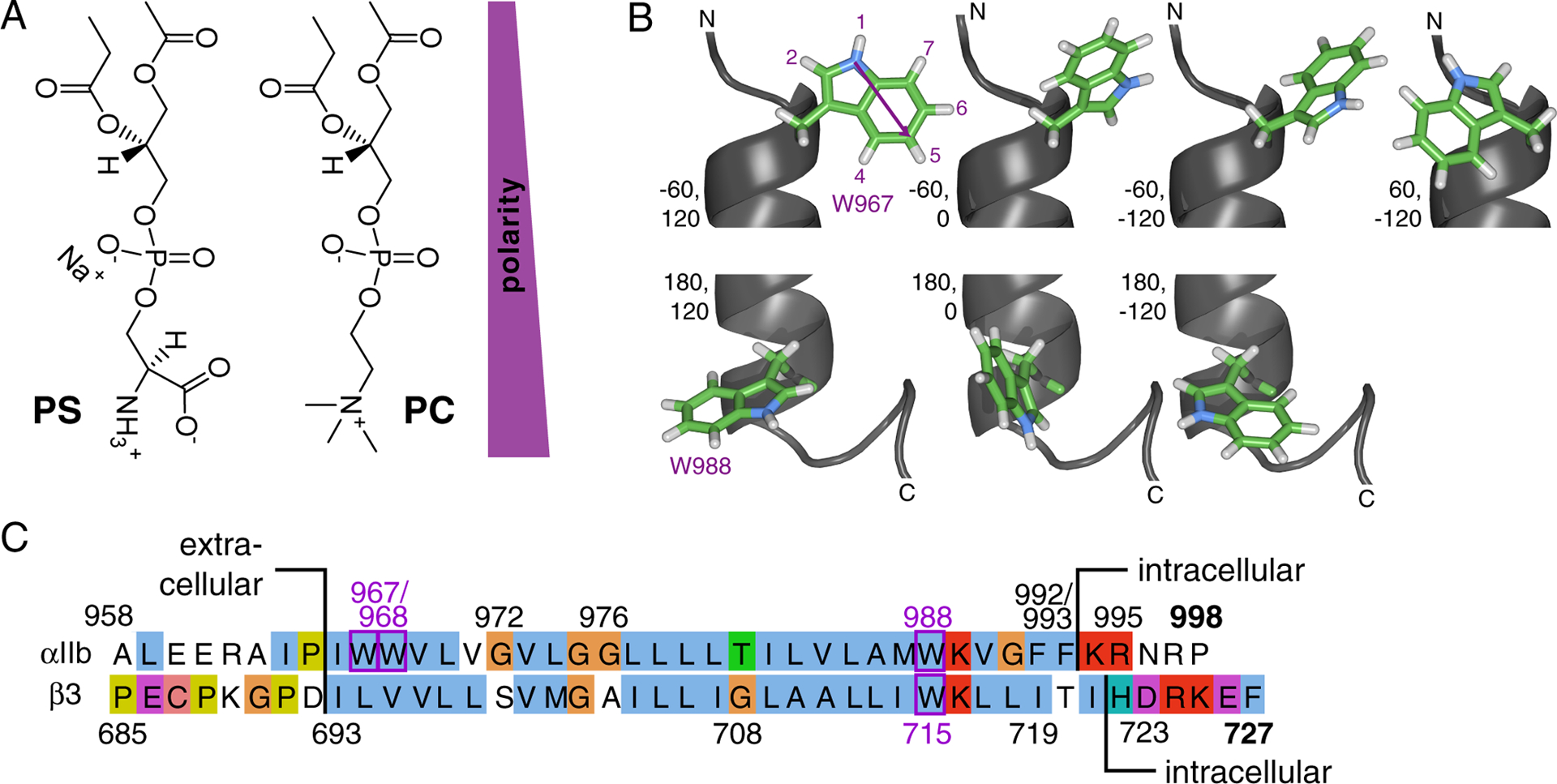
Overview of experimental system: lipids, tryptophan and integrin TM sequences. (A) Illustration of headgroup structures of phosphatidylserine (PS) and -choline (PC)-based lipids in extended conformations. (B) Structure of the Trp sidechain and rotamers populated.54 The indole ring has a permanent dipole moment of 2.1 Debye pointing from N1 in the five-membered ring to C5 in the six-membered ring.55 The N1 proton is the most acidic proton on indole and a hydrogen bond donor. Depicted are N-terminal W967 (χ1 ≈ −60°) and C-terminal W988 (χ1 ≈ 180°) of the integrin αIIb TM helix (PDB ID 2k1a)26 with χ2 ≈ −120°, 0°, 120°. For the benefit of completeness, W967 is also depicted with χ1 ≈ 60°, χ2 ≈ −120°. Rotamers were built using chimera.56 (C) Amino acid sequence of human integrin αIIb and β3 TM segments and flanking regions.
