Abstract
Purpose:
Quantitative in vivo molecular imaging of fine brain structures requires high-spatial resolution and high-sensitivity. Positron emission tomography (PET) is an attractive candidate to introduce molecular imaging into standard clinical care due to its highly targeted and versatile imaging capabilities based on the radiotracer being used. However, PET suffers from relatively poor spatial resolution compared to other clinical imaging modalities, which limits its ability to accurately quantify radiotracer uptake in brain regions and nuclei smaller than 3 mm in diameter. Here we introduce a new practical and cost-effective high-resolution and high-sensitivity brain-dedicated PET scanner, using our depth-encoding Prism-PET detector modules arranged in a conformal decagon geometry, to substantially reduce the partial volume effect and enable accurate radiotracer uptake quantification in small subcortical nuclei.
Methods:
Two Prism-PET brain scanner setups were proposed based on our 4-to-1 and 9-to-1 coupling of scintillators to readout pixels using 1.5 × 1.5 × 20 mm3 and 0.987 × 0.987 × 20 mm3 crystal columns, respectively. Monte Carlo simulations of our Prism-PET scanners, Siemens Biograph Vision, and United Imaging EXPLORER were performed using Geant4 application for tomographic emission (GATE). National Electrical Manufacturers Association (NEMA) standard was followed for the evaluation of spatial resolution, sensitivity, and count-rate performance. An ultra-micro hot spot phantom was simulated for assessing image quality. A modified Zubal brain phantom was utilized for radiotracer imaging simulations of 5-HT1A receptors, which are abundant in the raphe nuclei (RN), and norepinephrine transporters, which are highly concentrated in the bilateral locus coeruleus (LC).
Results:
The Prism-PET brain scanner with 1.5 mm crystals is superior to that with 1 mm crystals as the former offers better depth-of-interaction (DOI) resolution, which is key to realizing compact and conformal PET scanner geometries. We achieved uniform 1.3 mm full-width-at-half-maximum (FWHM) spatial resolutions across the entire transaxial field-of-view (FOV), a NEMA sensitivity of 52.1 kcps/MBq, and a peak noise equivalent count rate (NECR) of 957.8 kcps at 25.2 kBq/mL using 450–650 keV energy window. Hot spot phantom results demonstrate that our scanner can resolve regions as small as 1.35 mm in diameter at both center and 10 cm away from the center of the transaixal FOV. Both 5-HT1A receptor and norepinephrine transporter brain simulations prove that our Prism-PET scanner enables accurate quantification of radiotracer uptake in small brain regions, with a 1.8-fold and 2.6-fold improvement in the dorsal RN as well as a 3.2-fold and 4.4-fold improvement in the bilateral LC compared to the Biograph Vision and EXPLORER, respectively.
Conclusions:
Based on our simulation results, the proposed high-resolution and high-sensitivity Prism-PET brain scanner is a promising cost-effective candidate to achieve quantitative molecular neuroimaging of small but important brain regions with PET clinically viable.
Keywords: positron emission tomography, depth-of-interaction, high spatial resolution, high sensitivity, monoaminergic nuclei, time-of-flight, Prism-PET, quantitative molecular imaging
1 |. INTRODUCTION
Positron emission tomography (PET) is a versatile imaging modality that employs radiotracers for in vivo, quantitative characterization of neuroreceptors and other targets of interest within the central nervous system (CNS).1–3 PET tracers for molecular imaging are chosen on the basis of selective avidity to molecular targets. For example, [18F]AV-1451 binds to tau protein, a major component of Alzheimer’s disease (AD) pathology,4 and [11C]raclopride targets the dopamine D2 receptor, a protein linked to schizophrenia and neurochemical dysfunction in obesity.5,6
The tracers [11C]WAY-100635 and [18F]MefWAY specifically bind 5-HT1A serotonin receptors in the CNS, which are present at high density in the serotonergic brainstem raphe nuclei (RN), a major source of brain serotonin.7–11 The dorsal raphe nucleus (DRN) has been implicated in a variety of neuropsychiatric disorders,12–15 including highly prevalent conditions such as major depressive disorder (MDD)16–19 and AD.20–25 MDD is a heterogeneous disease that is often misdiagnosed and mistreated due to a lack of conclusive etiology.7,26. Thus, the DRN is an attractive candidate to be used as an imaging target for diagnosis, treatment selection, and treatment response in MDD patients.27–29
Although PET plays a key role in treatment validation and discovery of pathophysiological processes of brain diseases, conventional whole-body PET systems are still limited by poor spatial resolution and sensitivity. Low spatial resolution limits the ability to visualize small irregular brain structures and could drastically degrade the quantitative accuracy due to the partial volume effect (PVE).30,31 On the other hand, insufficient sensitivity results in a low signal-to-noise ratio (SNR), which restricts the capability of performing high-resolution and dynamic imaging.32 Although several dedicated brain PET scanners have been developed for research33–37 as well as proof-of-principle simulations to assess their capabilities,38–40 these systems still suffer from trade-offs among cost, sensitivity, spatial resolution, and timing resolution.
Despite having nearly six times higher 5-HT1A receptor density compared to cortical gray matter based on quantitative autoradiography,41 published receptor-specific binding measurements in the raphe PET images are lower than expected.42,43 This is due to the spatial resolution limitations of PET (on the order of 3–6 mm44,45) resulting in unwanted inclusion of low binding white matter within the volumes of interest (VOIs).46 Partial volume correction (PVC) may be used to mitigate PVE, but the choice of PVC method has been shown to drastically influence PET quantification results.47–49 Therefore, successful PET imaging of the RN necessitates improvements in spatial resolution of the PET scanner to substantially reduce the PVE.50
The locus coeruleus (LC) is another clinically relevant, small subcortical target in posterior area of the pons of the brainstem that has been difficult to image with PET. The LC is a bilateral structure and serves as the primary site of norepinephrine synthesis for the CNS.51,52 The LC has been implicated in a variety of diseases, including anxiety,53 post-traumatic stress disorder,54 and AD.55–59 However, similar to the RN, the LC cannot be accurately imaged and quantified with PET due to PVE, resulting in lower observable radiotracer uptake than expected relative to larger brain regions (i.e., cortical gray matter).60,61 In addition to PVE-related limitations, the LC has poor detection sensitivity in PET imaging due to its axially off-center position in a standard clinical PET ring’s field-of-view (FOV).62,63 PET systems with high geometric efficiency, such as small-diameter brain-dedicated scanners, would have greater success when imaging off-center VOIs such as the LC. However, there are currently no commercial small diameter human brain PET scanners due to the lack of practical and cost-effective depth-encoding methods that are required to mitigate the spatial blurring parallax error (PE).64
In this paper, we introduce our cost-effective, high-resolution, and high-sensitivity conformal brain PET scanner using our patented Prism-PET detector technology.65–72 Prism-PET detector blocks utilize segmented prismatoid light-guide array for localized and enhanced light sharing and their salient features are: 4-to-1 and 9-to-1 coupling of scintillator crystals to readout pixels,65,66 single-ended depth encoding readout with sub-2-mm depth-of-interaction (DOI) resolution,69 improved time-of-flight (TOF) performance using DOI corrections,67,68 and accurate decomposition of multi-crystal events for Compton scatter recovery (Ref. 70 and supplementary section in Ref. 65). We evaluated the imaging performance of our Prism-PET brain scanner and compared it with state-of-art whole-body and total-body TOF-PET scanners by performing Monte Carlo simulations based on the National Electrical Manufacturers Association (NEMA) guidelines, as well as brain simulations with relative specific binding values in the DRN and LC nuclei based on postmortem autoradiographic studies.73–75
2 |. MATERIALS AND METHODS
2.1 |. Scanner geometry
We simulated four different TOF-PET scanners in Geant4 application for tomographic emission (GATE)76 for comparison: the total body United Imaging EXPLORER, whole-body Siemens Biograph Vision, and our proposed TOF-DOI PET brain scanners based on Prism-PET detector modules with 0.987 × 0.987 × 20 mm3 and 1.5 × 1.5 × 20 mm3 crystals (Figure 1).
FIGURE 1.
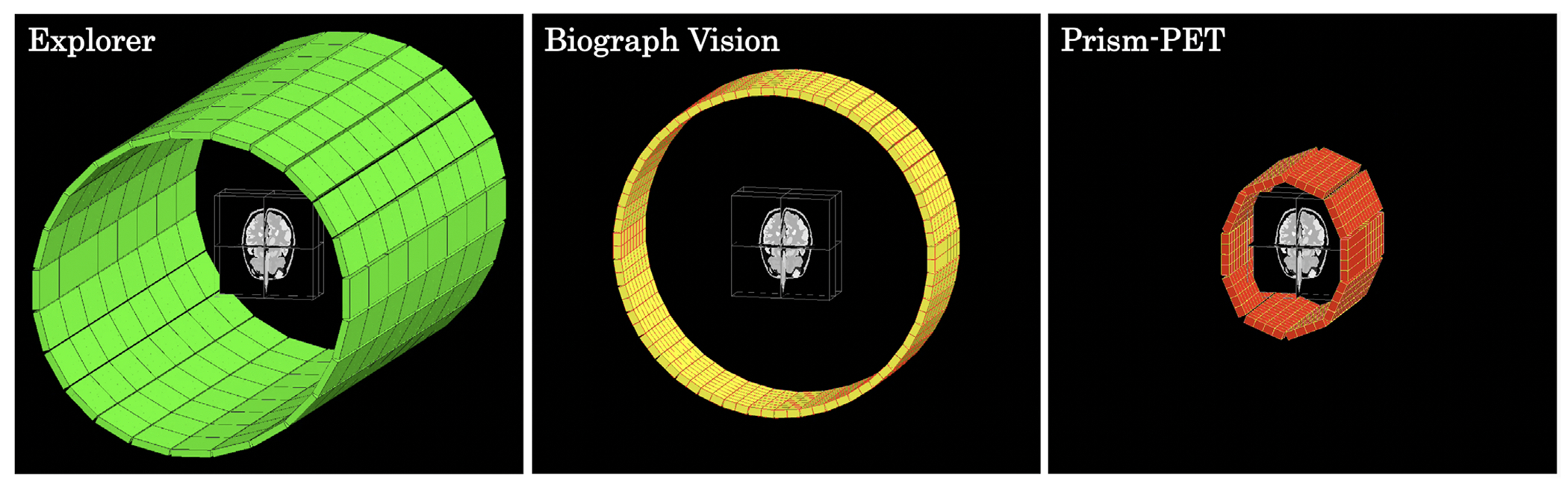
3D GATE models of the EXPLORER (left), Siemens Biograph Vision (middle), and Prism-PET brain scanner (right) with modified Zubal brain phantom at the center of the FOV.
The EXPLORER detector block consists of 2.76 × 2.76 × 18.1 mm3 lutetiumyttrium orthosilicate (LYSO) crystals in a 7 × 6 array (transaxial × axial) coupled to a 2 × 2 readout array of 6 × 6 mm2 silicon photomultiplier (SiPM) pixels. Seventy detector blocks of a 5 × 14 array form a detector module. Twenty-four corresponding detector modules form a section of the EXPLORER and eight sections in a row constitute the entire ~ 2 m axial length coverage with a 78.6 cm ring diameter, which enables PET imaging of the entire human body.77,78
The Biograph Vision detector block consists of a 20 × 10 array (transaxial × axial) of 3.2 × 3.2 × 20 mm3 lutetium oxyorthosillicate (LSO) ctystals. Each detector block is subdivided into eight mini-blocks which has 5 x 5 crystals coupled to a 4 × 4 array of SiPMs. The Biograph Vision consists of eight detector rings and 38 detector blocks per ring, forming a cylindrical scanner with 80 cm transaxial and 26.4 cm axial FOVs.45,79
Two Prism-PET brain scanner setups are proposed based on the single-ended 9-to-1 coupled and 4-to-1 coupled Prism-PET detector modules. The Prism-PET brain scanner with 1 mm crystals (Prism-PET 1 mm) was composed of 0.987 × 0.987 × 20 mm3 LYSO crystals arranged in 24 × 24 arrays coupled in ratios of 9-to-1 to 3 × 3 mm3 SiPM pixels.71 In comparison, the Prism-PET brain scanner with 1.5 mm crystals (Prism-PET 1.5 mm) was composed of 1.5 × 1.5 × 20 mm3 LYSO crystals arranged in 16 × 16 arrays coupled in ratios of 4-to-1 to 3 × 3 mm3 SiPM pixels. Crystal-to-SiPM ratios greater than one were used to simultaneously enhance spatial resolution, reduce readout cost, power consumption, and design complexity.65,66,72 To simultaneously increase geometrical sensitivity, reduce the spatial-blurring accolinearity, and further reduce cost, the Prism-PET scanners utilize an elliptical design that conforms to the human head with a small and large diameters of 29.1 and 38.5 cm, respectively. Both scanners consist of 14 detector rings and 40 detector blocks per ring, forming an axial FOV of 37.2 cm. Such a compact geometry is only possible thanks to the high DOI resolution of the Prism-PET modules.65,69
2.2 |. Performance evaluation
Simulations in this paper were performed using GATE and boosted by high-throughput computing (HTCondor) platform v8.8.9 running on five multithread work-stations each with 2 Intel-Xeon CPUs (44 threads/cpu) and 500 GB random-access memory. Detailed simulation parameters are listed in Table 1. Our simulation models of Biograph Vision and EXPLORER were validated by comparing the results of spatial resolution, system sensitivity, and count rate with previously published data.45,78,80 The imaging performance was assessed and quantified using the ultra-micro hot spot phantom and 3D voxelized brain phantoms.81
TABLE 1.
System configuration of the simulated scanners
| Parameters | EXPLORER | Biograph Vision | Prism 1 mm | Prism 1.5 mm |
|---|---|---|---|---|
| Scintillator materials | LYSO | LSO | LYSO | LYSO |
| Number of scintillators | 564,480 | 60,800 | 322,560 | 143,360 |
| Scintillator size (mm3) | 2.76 × 2.76 × 18.1 | 3.2 × 3.2 × 20 | 0.987 × 0.987 × 20 | 1.5 × 1.5 × 20 |
| Scintillator-to-SiPM ratio | 21:2 | 25:16 | 9:1 | 4:1 |
| Scintillator array dimension | 7 × 6 | 10 × 20 | 24 × 24 | 16 × 16 |
| Scintillators per ring | 840 | 760 | 960 | 640 |
| Number of rings | 672 | 80 | 336 | 224 |
| Axial length (mm) | 1941 | 264 | 372 | 372 |
| Long-ring diameter (mm) | 790 | 800 | 385 | 385 |
| Short-ring diameter (mm) | 790 | 800 | 290 | 290 |
| Coincidence time window (ns) | 6.0 | 4.7 | 2.5 | 2.5 |
| Energy window (keV) | 430–645 | 435–650 | 450–650 | 450–650 |
| DOI resolution (FWHM, mm) | N/A | N/A | 4.0 | 2.0 |
| TOF resolution (ps) | 500 | 200 | 250 | 200 |
Abbreviation: N/A, not applicable.
2.3 |. Spatial resolution
A 1 MBq 18F point source with a 0.3 mm diameter embedded in a 10 mm isotropic cube was used to evaluate spatial resolution of the four scanners. It was placed radially at 10, 25, 50, 75, and 100 mm from the center of the transaxial FOV and measurements were performed at axial positions of 0 mm and one-fourth the axial length away from the center of the scanner. At least 100K counts were collected in each measurement with the random coincidence rate being less than 5% of the total event rate.80
The inter-crystal scatter (ICS) recovery using the ground truth information from GATE was applied to both Prism-PET scanners to investigate the impact of Compton scatter on spatial resolution. The position of the first hit was used for the events that have Compton scattered inside the detector block. The ground truth DOI was calculated based on the distance from the hit position to the surface of the corresponding crystal. In order to accurately simulate the DOI performance of the Prism-PET detector modules, Gaussian noise with 4 and 2 mm full width at half maximum (FWHM) was added to the ground truth DOI of the Prism-PET 1 mm and 1.5 mm scanners, respectively.65,68,69
A virtual cylindrical scanner (VC) with 0.5 × 0.5 mm2 virtual crystals was modeled, shown as the yellow ring in Figure 2. Each list-mode event was rebinned to a pair of VC crystals along the same line of response (LOR). This procedure transferred the decagon geometry into a cylindrical one, which enabled the filtered backprojection (FBP) reconstruction using Software for Tomographic Image Reconstruction (STIR).82 Note that the VC rebinning was only used for the spatial resolution evaluation.
FIGURE 2.
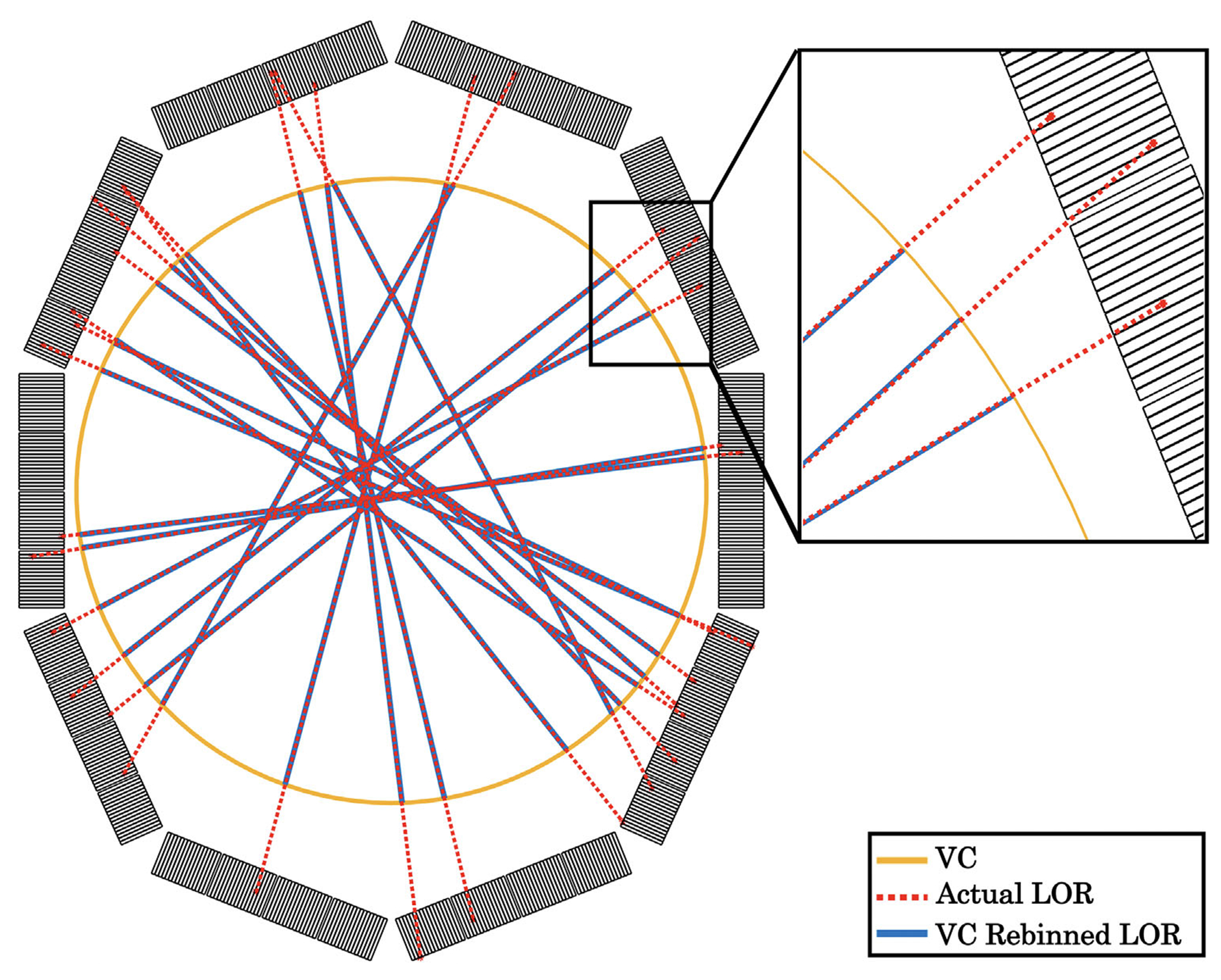
Demonstration of LORs rebinned to virtual cylinder (VC). Yellow: VC detector ring. Red dash line: actual LORs. Blue solid line: LORs rebinned to the VC
All list-mode data were reconstructed using the FBP 3D reprojection method (FBP-3DRP)82 with 0.3 × 0.3 × 0.25 mm3 voxel size for the Prism-PET 1 mm, 0.5 × 0.5 × 0.5 mm3 voxel size for Prism-PET 1.5 mm, 1.0 × 1.0 × 1.6 mm3 voxel size for Biograph Vision, and 1.0 × 1.0 × 1.42 mm3 voxel size for EXPLORER. All axial LORs were accepted for the Prism-PET brain scanners and Biograph Vision. A maximum ring difference of three axial sections (~72.6 cm) was set for the EXPLORER. No attenuation, scatter correction, or other post-processing was applied. The tangential, radial, and axial spatial resolution in both FWHM and full width at tenth maximum (FWTM) were determined according to NEMA NU 2-2012 guidelines.80
2.4 |. Axial sensitivity profile
A 700 mm polyethylene tube with a diameter of 1 mm was filled with 1 MBq of 18F and placed inside an 700 mm long aluminum sleeve with an inner diameter of 3.9 mm and an outer diameter of 6.4 mm. The phantom was placed at the center of the transaxial FOV and simulated for 100 s in GATE. The axial sensitivity profile was obtained by calculating the number of coincidences per 1.6 mm slice after single-slice rebinning (SSRB) and the counts were normalized by the phantom activity and scan time. System sensitivity was calculated by summing the count rate from all slices.80
2.5 |. Coincidence count rate
The coincidence count rate and noise equivalent count rate (NECR) were estimated following the NEMA NU 2-2012 guidelines. A polyethylene cylinder phantom with an outside diameter of 203 mm and an overall length of 700 mm was modeled as a scatter phantom. A 700 mm long line source was placed at a 45 mm radial distance from the central axis of the polyethylene phantom and filled with 0 to 1050 MBq 18F activity. Events that are 12 cm farther away from the center of transaxial FOV were excluded from the analysis. The NECR is given by , where the RTOT is the sum of true coincidence rate Rt, scattered coincidence rate Rs, and random coincidence rate Rr.80
2.6 |. Image quality
An ultra-micro hot spot phantom consisting of six rod groups with different diameters (2.4, 2.0, 1.7, 1.35, 1.0, and 0.75 mm) was used to evalute the image quality. The phantom was placed at both 0 mm and 100 mm from the center of the transaxial FOV. The rods were filled with 250 kBq/cc of 18F and the rest of the phantom was filled with water.
The ICS recovery was applied to the Prism-PET 1 and 1.5 mm scanners as described in the spatial resolution section. DOI-rebinning was performed at the list-mode level to achieve PE correction (the crystal pair with their radiation entrance surface intersecting the LOR were identified as the coincidence channels for each event).83 This procedure was applied in both transaxial and axial direction to achieve fully 3D PE correction. Images were reconstructed using open-source Customizable and Advanced Software for Tomographic Reconstruction (CASToR)84 with 3D list-mode ordered subset expectation maximization (OSEM) and point-spread function (PSF) modeling. The OSEM reconstruction parameters are listed in Table 2. No other post-processing was applied to the reconstructed images.
TABLE 2.
CASToR reconstruction parameters
| Scanner | Matrix size | Voxel size (mm) | PSF modelinga | Iteration subsets | Simulation time (s) |
|---|---|---|---|---|---|
| EXPLORER | 512 × 512 × 256 | 0.5 × 0.5 × 0.5 | 3.0:3.0 | 5:8 | 120 |
| Biograh Vision | 512 × 512 × 256 | 0.5 × 0.5 × 0.5 | 3.5:3.5 | 5:8 | 120 |
| Prism-PET 1 mm | 512 × 512 × 256 | 0.5 × 0.5 × 0.5 | 1.5:1.5 | 5:8 | 120 |
| Prism-PET 1.5 mm | 512 × 512 × 256 | 0.5 × 0.5 × 0.5 | 1.5:1.5 | 5:8 | 120 |
The PSF is modeled as a 3D Gaussian kernel with transaxial-FWHM : axial-FWHM in mm.
2.7 |. Brain simulation
We simulated the 3D voxelized Zubal human brain phantom with high-resolution parcellation (1 mm isotropic voxels and 256 × 256 × 128 matrix size) based on MRI and CT data of two healthy male adults.81 Three VOIs were added to the Zubal phantom: DRN, left locus coeruleus (LLC), and right locus coeruleus (RLC). The DRN was added along the midline of the brainstem, while the two LC VOIs were symmetrically placed in the brainstem adjacent to the fourth ventricle based on coordinates provided in the literature.85–88 The DRN was inserted as a 3-mm diameter sphere,73,85 while both the LLC and RLC were modeled as 2-mm diameter spheres with their centers placed 2 mm lateral to the midline of the brain.63,89
For each of the four simulated scanners, two separate simulations were performed: one based on relative uptake of [11C]WAY-100635, a 5-HT1A receptor radiotracer,73,74 and one based on relative uptake of (S,S)-[18F]FMeNER-D2, a norepinephrine transporter (NET) radiotracer.75 Total activities in the GATE simulations were 0.16 mCi and 0.2 mCi for the imaging studies of the seretonin receptor and NET, respectively. Note that while these doses are much lower than what’s typically administered in clinical PET scans, they are simulated exclusively in the brain whereas in real scans radiotracers generally have much higher nonspecific binding and will be distributed to other regions and even throughout the entire body. In addition, the simulated tracers have high specificity for the brain regions being studied with high signal-to-background ratios, and thus high-quality images with sufficient contrast can be acquired at low doses. Uptake in each brain region is primarily based on the radiopharmaceutical rather than the radioisotope attached to it. For example, many analogs of WAY-100635 have been developed that have 18F as the radioisotope, including [18F]FCWAY90 and [18F]Mefway,91 which provide better intrinsic spatial resolution due to the lower positron range of 18F compared to 11C.44
Each voxel of the 3D brain phantom was attached with 18F activity based on autoradiographic studies showing the specific binding of 5-HT1A radiotracer73,74 and NET75 relative to the DRN and LC, respectively (Table 3). Radioactive decay of the 18F source, positron range of 18F, and photon pair acollinearity were all modeled in GATE. The coincidence windows used for the EXPLORER, Biograph Vision, and Prism-PET scanners were 6, 4.73, and 2.5 ns, respectively. The scanners’ temporal resolution was simulated by setting the single crystal’s time-domain Gaussian blurring parameter (FWHM) in GATE where coincidence time resolution (CTR) is the quadrature sum of two coincidence crystals’ timing resolutions. The CTR of each scanner is listed in the TOF resolution section in Table 1.
TABLE 3.
Specific binding values used in the 5-HT1A receptor and NET PET simulations
| Brain regions | Specific binding relative to DRN73,74 | Brain regions | Specific binding relative to LC75 |
|---|---|---|---|
| DRN | 100.0 | LC | 100.0 |
| Hippocampus | 74.0 | Pons anterior | 9.8 |
| Uncus | 77.0 | Caudatus | 5.7 |
| Amygdala | 10.0 | Putamen | 3.0 |
| Entorhinal cortex | 44.0 | Thalamus | 8.0 |
| Insular cortex | 25.0 | Temporal cortex | 12.3 |
| Temporal polar cortex | 43.0 | Insular cortex | 9.9 |
| Frontal cortex | 18.0 | Occipital cortex | 1.6 |
| Temporal cortex | 22.0 | Cerebellum, total | 15.2 |
| Occipital cortex | 10.0 | Cerebellum, gray matter | 15.6 |
| Caudate nucleus | 1.0 | White matter | 7.6 |
| Globus pallidus | 1.0 | N/A | N/A |
| Putamen | 1.0 | N/A | N/A |
| Thalamus | 2.0 | N/A | N/A |
Abbreviations: NET, norepinephrine transporter; PET, positron emission tomography; DRN, dorsal raphe nucleus; LC, locus coeruleus; N/A, not applicable.
The list-mode data of all axial LORs was utilized while those for the Prism-PET brain scanners were further processed for ICS recovery and DOI-rebinning (see the spatial resolution and image quality sections). The images were reconstructed by CASToR using 3D list-mode TOF-OSEM algorithm with PSF modeling (see Table 2).84 No scatter correction was performed for the PET images but the attenuation correction was applied by using an attenuation map generated from the Zubal brain phantom where tissues were segmented into four groups: bone, air, blood, and soft tissue.
Reconstructed brain simulation images were registered to the modified Zubal phantom using a six-parameter rigid-body transform in SPM1292 for quantitative analysis. We calculated the average voxel intensity, which is a quantitative metric representative of volumetric uptake, of each of the three added nuclei VOIs in order to characterize the capabilities of the simulated scanners in resolving small brain structures. The VOIs used for signal extraction were identical to the ones that were inserted into the brain for the GATE simulations (i.e., 3 mm sphere for the DRN and 2 mm spheres for the LLC and RLC). To quantitatively characterize contrast, we calculated the relative uptake ratio (denoted RUR) as the average voxel intensity of the VOI versus that of the entire cortical gray matter, the latter being our reference region.
3 |. RESULTS
3.1 |. Spatial resolution, coincidence count rate, and sensitivity
Table 4 shows the spatial resolutions in FWHM and FWTM at 1, 5, and 10 cm from the center of the transaxial FOV. The system sensitivity values and peak NECRs are also summarized in Table 4 with the axial sensitivity profile shown in Figure 3. The Prism-PET scanners achieved more than three times higher system sensitivity compared to the Biograph Vision. The Prism-PET 1 mm has a slightly lower sensitivity than Prism-PET 1.5 mm because of it’s smaller crystal volume. Figure 4 depicts the trues, scatters, randoms, total (prompt) count rates, and NECRs of the four scanners as a function of activity. The maximum NECRs are 1367.4 kcps at 16.8 kBq/cc for the EXPLORER,~950 kcps at 25.2 kBq/cc for the Prism-PET scanners, and 290 kcps at 25.2 kBq/cc for the Biograph Vision.
TABLE 4.
Evaluation results of the simulated scanners in GATE based on NEMA guidelines
| Parameters | Distancea | EXPLORER | Biograph Vision | Prism-PET 1 mmb | Prism-PET 1.5 mmb | ||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| Spatial resolution (mm) | FWHM | FWTM | FWHM | FWTM | FWHM | FWTM | FWHM | FWTM | |
| Radial | 1 | 2.92 | 5.33 | 3.30 | 6.04 | 1.23 | 2.24 | 1.22 | 2.22 |
| 5 | 3.10 | 5.66 | 3.34 | 6.11 | 1.25 | 2.28 | 1.20 | 2.18 | |
| 10 | 4.49 | 8.20 | 3.76 | 6.86 | 1.54 | 2.80 | 1.26 | 2.29 | |
| Tangential | 1 | 2.83 | 5.17 | 3.34 | 6.10 | 1.21 | 2.20 | 1.33 | 2.42 |
| 5 | 4.23 | 7.72 | 3.28 | 5.98 | 1.31 | 2.38 | 1.43 | 2.60 | |
| 10 | 4.13 | 7.54 | 3.50 | 6.39 | 1.29 | 2.35 | 1.33 | 2.42 | |
| Axial | 1 | 4.26 | 7.78 | 4.43 | 8.09 | 1.16 | 2.11 | 1.26 | 2.34 |
| 5 | 4.30 | 7.85 | 4.47 | 8.16 | 1.19 | 2.17 | 1.31 | 2.30 | |
| 10 | 5.27 | 9.62 | 4.59 | 8.38 | 1.17 | 2.13 | 1.29 | 2.35 | |
| Sensitivity (kcps/MBq) | 197.9 | 16.3 | 50.4 | 52.1 | |||||
| Peak NECR (kcps at kBq/mL) | 1367.4 at 16.8 | 290.0 at 25.2 | 923.6 at 25.2 | 957.8 at 25.2 | |||||
Abbreviations: DOI, depth-of-interaction; FOV, field of view; FWHM, full width at half maximum; FWTM, full width at tenth maximum; GATE, Geant4 application for tomographic emission; NECR, noise equivalent count rate; NEMA, National Electrical Manufacturers Association.
Distance (in cm) from the center of transaxial FOV.
ICS + DOI corrections were applied to the Prism-PET scanners.
FIGURE 3.
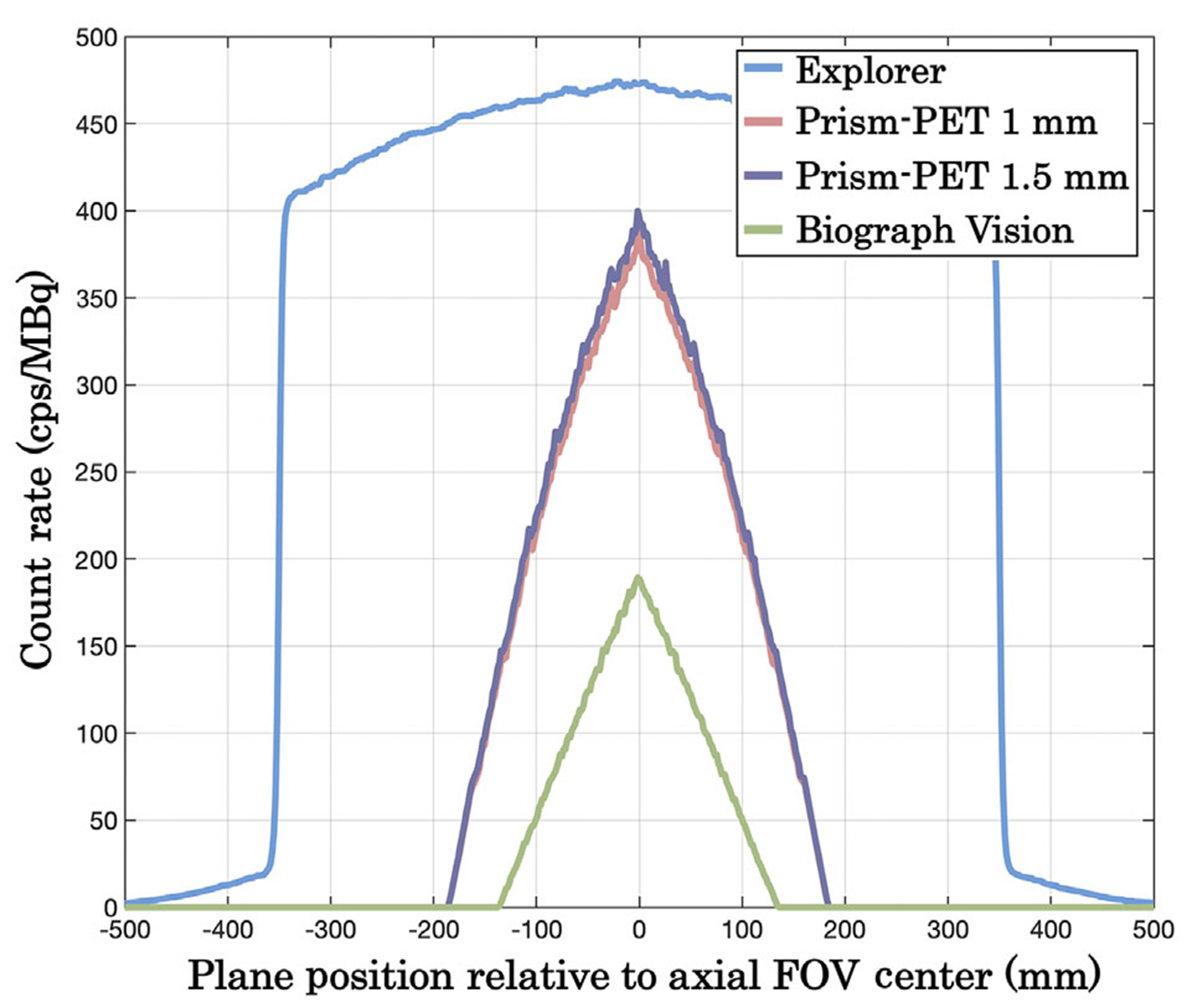
Axial sensitivity profiles of EXPLORER, Biograph Vision, Prism-PET 1 mm, and Prism-PET 1.5 mm.
FIGURE 4.
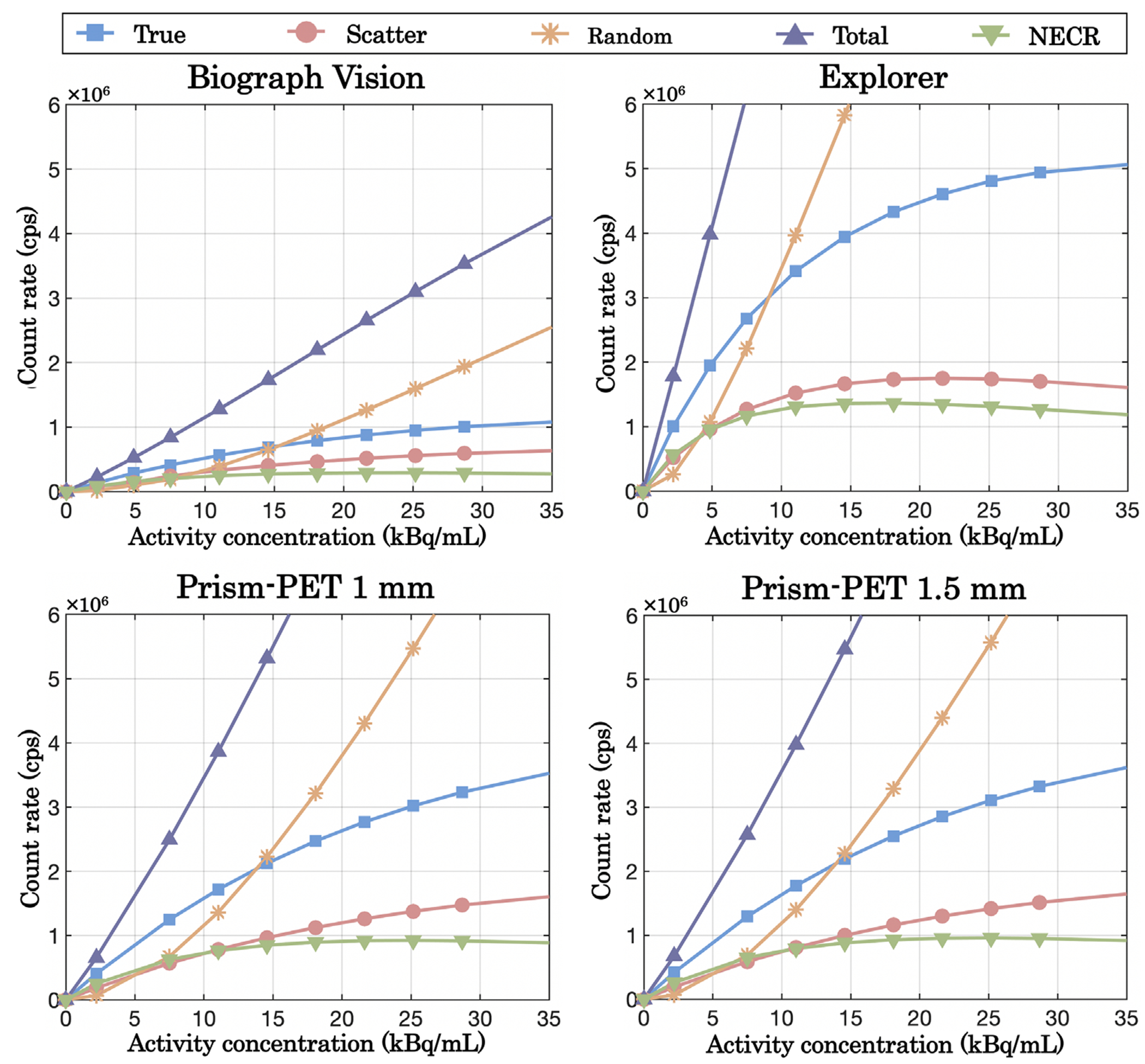
Count rate and NECR performance of EXPLORER, Biograph Vision, Prism-PET 1 mm, and Prism-PET 1.5 mm.
3.2 |. Image quality and 3D voxelized brain phantom
The ultra-micro hot spot phantom images are shown in Figure 5. All 1.35 mm hot spots can be distinctly resolved at both the center and edge of the FOV by both Prism-PET scanners after the ICS recovery and DOI rebinning, while the Biograph Vision and EXPLORER can only resolve the largest hot spots with 2.4 mm in diameter. The spatial blurring of Compton scatter can clearly be seen for ultra-high resolution PET imaging after DOI correction and while comparing the reconstructed images without and with ICS recovery.
FIGURE 5.
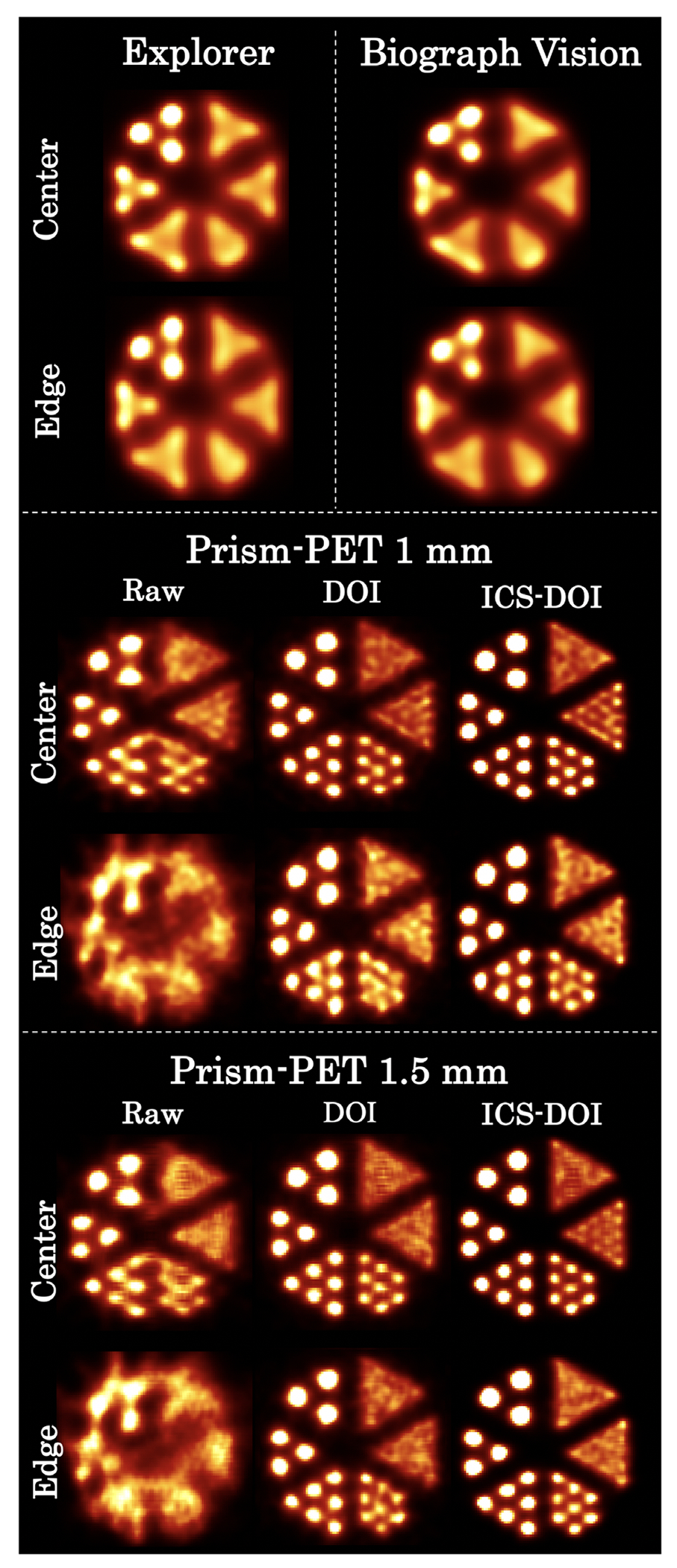
Simulated ultra-micro phantom images at the center and edge (i.e., 100 mm offset from the center of the transaxial FOV). Rods’ diameters from smallest to largest are 0.75, 1.0, 1.35, 1.7, 2.0, and 2.4 mm (clockwise). Raw: images without any corrections. DOI: images with DOI-rebinning. ICS-DOI: images with both ICS recovery and DOI-rebinning.
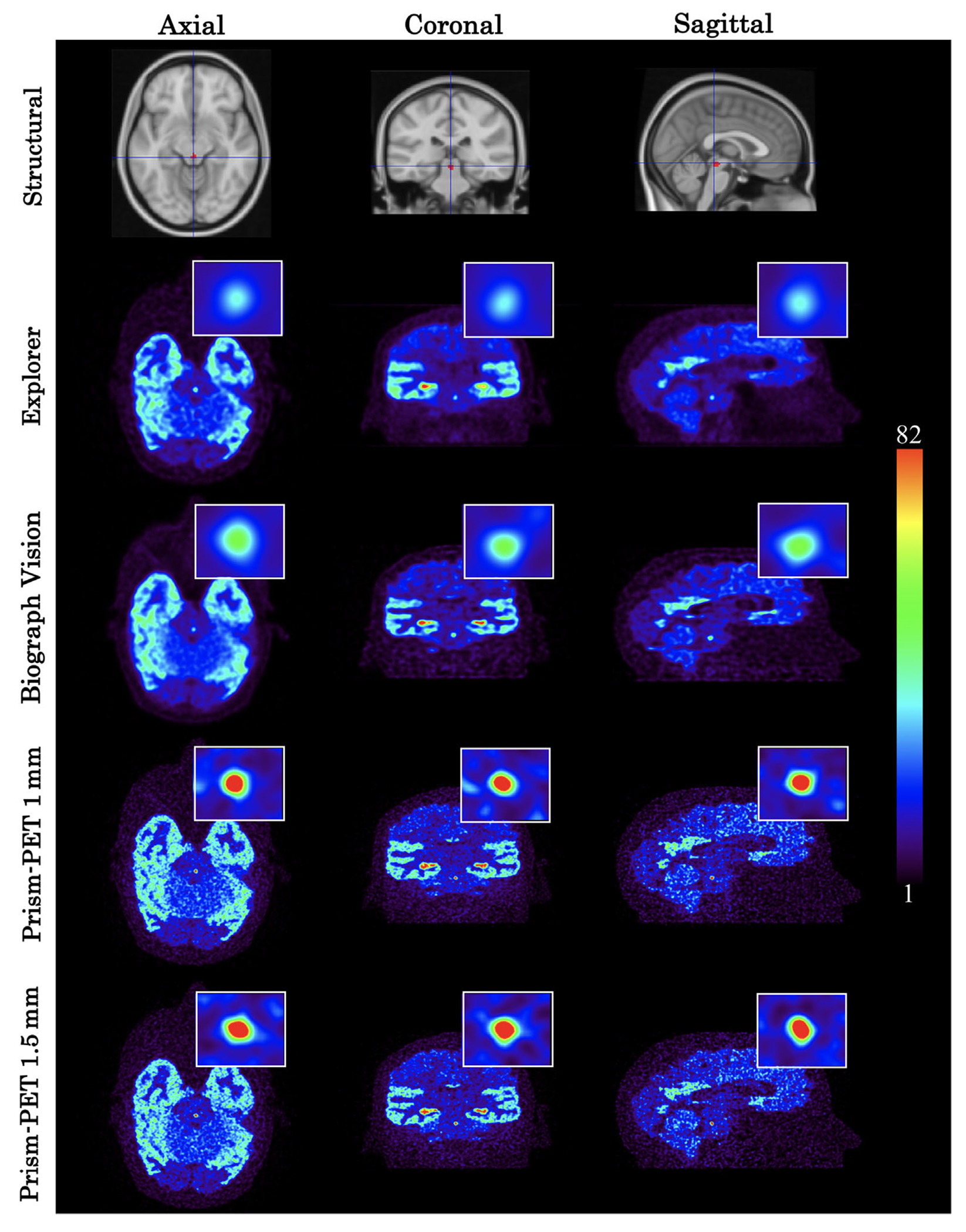
The Prism-PET scanners demonstrated more accurate radiotracer uptake when imaging the 5-HT1A receptor and NET than both the Biograph Vision and EXPLORER scanners relative to the uptake in the entire cortical gray matter (Figures 6 and 7). In the serotonin PET study, the Prism-PET scanners enabled high-contrast visualization of the DRN, while neither the Biograph Vision’s nor the EXPLORER’s spatial resolution permitted clear distinction of the DRN (Figure 6). The relative uptake ratio between the DRN and cortical gray matter was much higher in the Prism-PET scanners (Prism-PET 1 mm: RURDRN = 3.3; Prism-PET 1.5 mm: RURDRN = 3.5) compared to both the Biograph Vision (RURDRN = 1.9) and EXPLORER (RURDRN = 1.3), representing a 1.8-fold and 2.6-fold improvement in quantification accuracy, respectively (Figure 8). In addition, the hippocampal head is clearly visible in the Prism-PET scanner along with the subiculum due to the enhanced spatial resolution, whereas the substructures of the hippocampus cannot be distinguished with the Biograph Vision or EXPLORER (Figure 6).
FIGURE 6.
5-HT1A radiotracer GATE simulation using the Biograph Vision, EXPLORER, Prism-PET 1 mm, and Prism-PET 1.5 mm. The insets in these figures show a zoomed-in view of the DRN. Top row represents the corresponding slice in Montreal Neurological Institute (MNI152) space (centered around the DRN).
FIGURE 7.
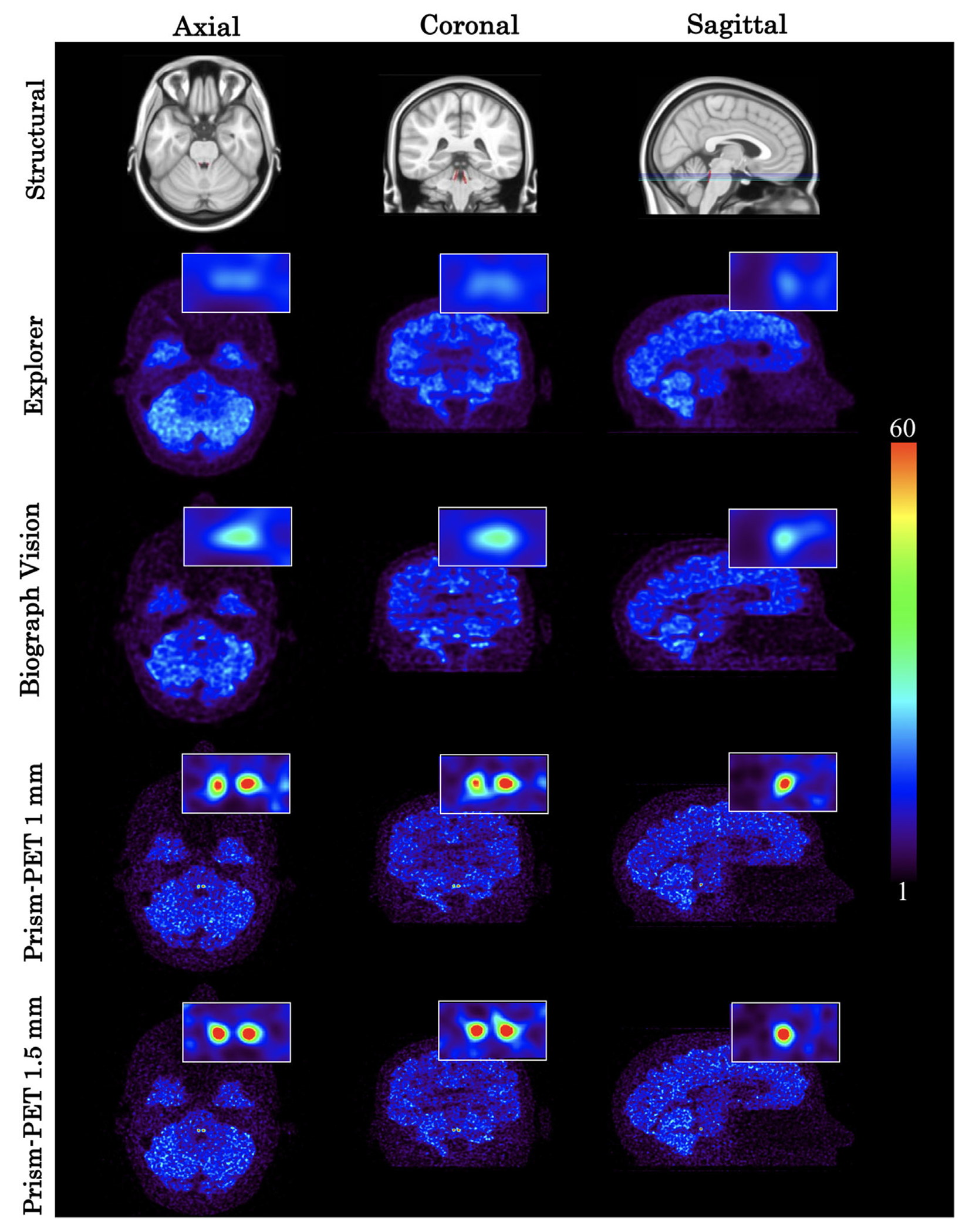
NET radiotracer GATE simulation using the EXPLORER, Biograph Vision, Prism-PET 1 mm, and Prism-PET 1.5 mm. The insets in these figures show a zoomed-in view of the bilateral LC. Top row represents the corresponding slice in MNI152 template images (centered around the LC).
FIGURE 8.
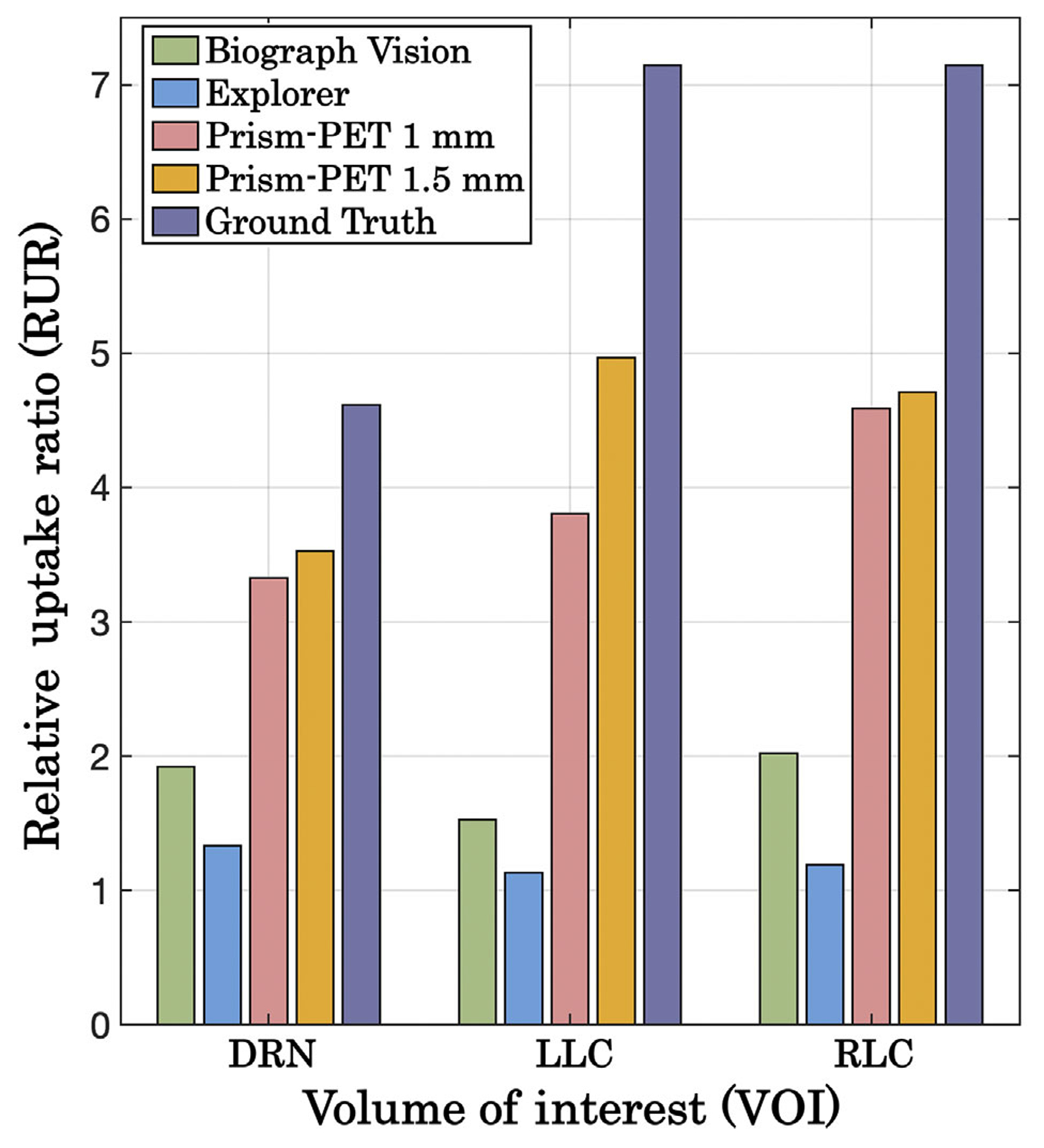
Calculated RUR values for each VOI (DRN, LLC, and RLC) using the 5-HT1A radiotracer (DRN) and NET radiotracer (LLC, RLC) for all four simulated scanners. Ground truth RUR values based on postmortem autoradiographic studies are also displayed for reference.
In the NET PET study, the LLC and RLC were not distinguishable in the Biograph Vision and EXPLORER images, while the Prism-PET scanners allowed for clear visualization and distinction of each VOI (Figure 7). Similar to the serotonin receptor quantitation, the relative uptake ratios between the LC VOIs and the cortex were noticeably higher using the Prism-PET scanners (Prism-PET 1 mm: RURLLC = 3.8, RURRLC = 4.6; Prism-PET 1.5 mm: RURLLC = 5.0, RURRLC = 4.7) compared to the Biograph Vision (RURLLC = 1.5, RURRLC = 2.0) and EXPLORER (RURLLC = 1.1, RURRLC = 1.2, see Figure 8).
4 |. DISCUSSION
The decagon-shaped geometry of the Prism-PET scanners (1) provides a larger solid angle coverage which improves sensitivity (52.1 kcps/MBq for Prism-PET 1.5 mm vs. 16.3 kcps/MBq for Biograph Vision), (2) reduces the spatial blurring acollinearity as the detector blocks closely wrap around the human head for substantially smaller LOR lengths, and (3) reduces the crystal surface area (a significant cost factor of a PET scanner) by 50% and 92% compared to the Biograph Vision and EXPLORER, respectively. However, since the conformal decagon geometry increases the fraction of gamma photons that hit the crystal surface with oblique angles, Prism-PET brain scanners suffer from a high degree of PE, leading to the degradation of spatial resolution even at the center of the FOV, as well as severe blurring towards the edge of the FOV.44 The parallax artifact can be mitigated by enhancing the positional accuracy of the LORs using events’ DOI information.93 By applying DOI-rebinning,83 the parallax artifact in both the transaxial and axial FOV is corrected and a substantially higher image quality is obtained across the entire FOV (see Figure 5). Although the Prism-PET 1.5 mm (with 2 mm DOI resolution) uses larger crystal elements, it provided better spatial resolution than the Prism-PET 1 mm (with 4 mm DOI resolution) especially for regions near the edge of the transaxial FOV, suggesting that accurate DOI localization is indispensable for compact and conformal PET scanners to achieve uniform high spatial resolution. One important consideration is that the number of LORs increases quadratically with the number of DOI layers which may lead to higher computational costs for DOI-based image reconstruction.94,95 One solution is to perform DOI-rebinning in list-mode followed by a conventional non-DOI image reconstruction. Thus, although continuous DOI was simulated for the Prism-PET scanners, the image reconstruction time remained similar to that of non-DOI scanners.
Compton scatter is another factor that further exacerbates the trade-off between spatial resolution and sensitivity unless the position of the first interaction site is recovered.96 For small crystals used by Prism-PET scanners, a large percentage of 511 keV photons will scatter to the adjacent crystals and cause mispositioning of the LORs which leads to spatial blurring and image quality degradation.97,98 Although some of the scattered events are rejected due to insufficient energy deposition and/or collection, the majority of them are detected to boost sensitivity and image SNR. One of the major advantages of the Prism-PET detector block is the enhanced and localized light-sharing of scintillation photons to neighboring SiPM pixels with a characteristic pattern (supplementary section in Ref. 65). This enables accurate decomposition of energies and DOIs for both the scattered photon and the recoil electron in the multicrystal event. The knowledge of energy and DOI for interactions in each crystal results in the most accurate ICS recovery70,99 and substantially mitigates the trade-off between sensitivity and spatial resolution when including ICS events in the image reconstruction. The hot spot phantom simulation results validate that the Prism-PET’s ICS recovery can enhance spatial resolution and the ability to resolve and quantify uptake in small structures (Figure 5).
Quantitative PET imaging for small brain structures relies on having sufficient sensitivity and spatial resolution in order to accurately assess radiotracer uptake with minimal PVE.50,100 While the current state-of-the-art Biograph Vision and total-body EXPLORER scanners cannot accurately visualize and quantify uptakes in the LLC, RLC, and DRN (RUR< 2 for all three VOIs in both scanners), our proposed depth encoding Prism-PET scanners offer higher RUR values in all three VOIs due to about an order-of-magnitude higher volumetric resolution and substantially lower PVE (Figures 6–8).65,66 One must note that only the Prism-PET 1.5 mm scanner obtained bilateral and symmetrical uptakes with maximum RURs in the LLC and RLC nuclei thanks to its superior DOI resolution which yields the most accurate positioning of LORs and least PE.
The high-resolution research tomograph (HRRT),101 which is designed over 20 years ago and is no longer commercially available, is the most widely used brain PET scanner and serves as a performance benchmark for new high-resolution systems. However, HRRT’s moderate spatial resolution (2.4 mm at the center of the FOV), insufficient DOI localization accuracy (two-layer discrete DOI) given the compact octagon geometry, poor sensitivity (4.3% at the center of FOV), and absence of TOF readout have substantially limited its quantitative accuracy for the numerous studies performed at the 17 installed sites around the world.102 For example, low SNR and severe PVE in the HRRT [11C]MRB-PET images contributed to the lowest measured NET concentration in the LC region.103 In another recent study, small regions such as the raphe nuclei were excluded from any quantitative analysis because PVE led to a severe underestimation of binding potentials in small and high binding regions (i.e., raphe) which are surrounded by low binding tissue (i.e., white matter tissue of the brainstem).42
Because of the trade-off between sensitivity and spatial resolution for large axial FOV non-DOI PET scanners, we observe that although the EXPLORER has the highest sensitivity and NECR, it has the lowest quantitative accuracy in both the 5-HT1A receptor and NET PET studies. The axial detector penetration of obliquely incident gamma photons detected within the wide acceptance angle in the EXPLORER introduces significant PE which leads to degraded spatial resolution, as shown in a recent simulation study.104 This axial PE can be reduced by limiting the accepted ring difference at the expense of reduced sensitivity.105 In addition, the EXPLORER has a poor TOF resolution of ~ 500 ps, which together with the absence of depth-encoding offset its effective sensitivity gain, especially for imaging single organs such as the human brain.106
Accurate and reliable imaging of the molecular and functional attributes of the RN and LC could lead to a better understanding of brain physiology and pathology (e.g., AD pathogenesis), as well as assist with clinical decision making. Accumulation of tau and neuronal loss has been observed in the cortical and subcortical brain sites as AD progresses.107–109 Studies suggest that small subcortical brain regions are involved in early AD before cytoskeletal changes occur in the entorhinal cortex, and the LC and RN are the first affected structures.110,111 However, the application of, for example, LC FDG-PET imaging as a potential in vivo biomarker of AD is not supported because of significant PVE in the acquired patient data.112 Our high-resolution Prism-PET brain scanner enables quantitative imaging of LC and other small brain nuclei which may lead to alternative findings and help discover potential imaging biomarkers of the AD. In addition, observing changes in the LC and RN in response to lifestyle adjustments or medical therapy could also guide the discovery and application of new and effective treatments. The molecular imaging of LC and RN may offer a more sensitive and specific addition to the cognitive, functional, and behavioral batteries that are currently used for diagnosis and monitoring disorders of the CNS.
5 |. CONCLUSION
A next-generation TOF-DOI brain PET scanner is proposed using our recently developed Prism-PET detector technologies to simultaneously and cost-effectively achieve high resolution and high sensitivity. Performance evaluations using NEMA guidelines and ultra-micro hot spot phantom simulations demonstrated a system sensitivity of 52.1 kcps/MBq while achieving 1.3 mm spatial resolution across the entire FOV, thanks to (1) 4-to-1 crystal-to-pixel coupling (using 1.5 mm crystals and 3 mm SiPM pixels), (2) single-ended depth encoding and TOF readout (with 2 mm FWHM DOI resolution), (3) compact and conformal scanner geometry (reducing spatial blur due to accolinearity with a twofold reduction in scintillator volume compared to the Biograph Vision), and (4) corrections of ICS events. The 3D voxelized brain phantom simulations showed that the Prism-PET brain scanner, with substantially reduced PVE and improved SNR, enables accurate visualization and uptake quantification of 5-HT1A radiotracer in the DRN and NET radiotracer in the LC. The ultimate goal is to provide ultra-high performance PET neuroimaging at reduced cost for increased geographical and clinical dissemination and also to have a significant clinical impact by enabling high SNR and high-resolution parametric imaging with voxel-level kinetic modeling and reliable quantitative in vivo measurement of molecular targets in a host of small brain nuclei.
ACKNOWLEDGMENTS
The authors would like to thank Center for Biotechnology at Stony Brook University for their financial support through the NIH REACH program. The authors also thank Thibaut Merlin and Simon Stute for their assistance with the CASToR software. Research reported in this work was supported by NIH under award number U01 HL127522 and grant No. R01 EB030413.
Footnotes
CONFLICT OF INTEREST
The authors have declared no conflict of interest.
REFERENCES
- 1.Laruelle M. Imaging synaptic neurotransmission with in vivo binding competition techniques: a critical review. J Cereb Blood Flow Metab. 2000;20:423–451. [DOI] [PubMed] [Google Scholar]
- 2.Nasrallah I, Dubroff J.An overview of PET neuroimaging.Semin Nucl Med. 2013;43:449–461. [DOI] [PubMed] [Google Scholar]
- 3.Paterson LM, Kornum BR, Nutt DJ, Pike VW, Knudsen GM. 5-HT radioligands for human brain imaging with PET and SPECT. Med Res Rev. 2013;33:54–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mishra S, Gordon BA, Su Y, et al. AV-1451 PET imaging of tau pathology in preclinical Alzheimer disease: defining a summary measure. NeuroImage. 2017;161:171–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farde L, Hall H, Ehrin E, Sedvall G. Quantitative analysis of D2 dopamine receptor binding in the living human brain by PET. Science. 1986;231:258–261. [DOI] [PubMed] [Google Scholar]
- 6.Wang G-J, Volkow ND, Logan J, et al. Brain dopamine and obesity. The Lancet. 2001;357:354–357. [DOI] [PubMed] [Google Scholar]
- 7.Pillai RL, Zhang M, Yang J, et al. Will imaging individual raphe nuclei in males with major depressive disorder enhance diagnostic sensitivity and specificity? Depress Anxiety. 2018;35:411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parsey RV, Oquendo MA, Simpson NR, et al. Effects of sex, age, aggressive traits in man on brain serotonin 5-HT1A receptor binding potential measured by PET using [C-11]WAY-100635.Brain Res. 2002;954:173–182. [DOI] [PubMed] [Google Scholar]
- 9.Saigal N, Pichika R, Easwaramoorthy B, et al. Synthesis and biologic evaluation of a novel serotonin 5-HT1A receptor radioligand, 18F-labeled mefway, in rodents and imaging by PET in a nonhuman primate. J Nucl Med. 2006;47:1697–1706. [PubMed] [Google Scholar]
- 10.Hillmer AT, Wooten DW, Bajwa AK, et al. First-in-human evaluation of 18F-mefway, a PET radioligand specific to serotonin-1A receptors. J Nucl Med. 2014;55:1973–1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pike VW, McCarron JA, Lammerstma AA, et al. First delineation of 5-HT1A receptors in human brain with PET and [11C]WAY-100635. Eur J Pharmacol. 1995;283:R1–R3. [DOI] [PubMed] [Google Scholar]
- 12.Sullivan GM, Ogden RT, Oquendo MA, et al. Positron emission tomography quantification of serotonin-1A receptor binding in medication-free bipolar depression. Biol Psychiatry. 2009;66:223–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Toczek MT, Carson RE, Lang L, et al. PET imaging of 5-HT1A receptor binding in patients with temporal lobe epilepsy. Neurology. 2003;60:749–756. [DOI] [PubMed] [Google Scholar]
- 14.Savic I, Lindström P, Gulyás B, Halldin C, Andrée B, Farde L. Limbic reductions of 5-HT1A receptor binding in human temporal lobe epilepsy. Neurology. 2004;62:1343–1351. [DOI] [PubMed] [Google Scholar]
- 15.Drevets WC, Frank E, Price JC, et al. Pet imaging of serotonin 1A receptor binding in depression. Biol Psychiatry. 1999;46:1375–1387. [DOI] [PubMed] [Google Scholar]
- 16.Lira A, Zhou M, Castanon N, et al. Altered depression-related behaviors and functional changes in the dorsal raphe nucleus of serotonin transporter-deficient mice. Biol Psychiatry. 2003;54:960–971. [DOI] [PubMed] [Google Scholar]
- 17.Boldrini M, Underwood MD, Mann JJ, Arango V. Serotonin-1A autoreceptor binding in the dorsal raphe nucleus of depressed suicides. J Psychol Res. 2008;42:433–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Michelsen KA, Schmitz C, Steinbusch HW. The dorsal raphe nucleus-From silver stainings to a role in depression. Brain Res Rev. 2007;55:329–342. [DOI] [PubMed] [Google Scholar]
- 19.Wohlschläger A, Karne H, Jordan D, Lowe MJ, Jones SE, Anand A. Spectral dynamics of resting state fMRI within the ventral tegmental area and dorsal raphe nuclei in medication-free major depressive disorder in young adults. Front Psychiatry. 2018;9:163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stratmann K, Heinsen H, Korf H-W, et al. Precortical phase of Alzheimer’s disease (AD)-related tau cytoskeletal pathology. Brain Pathol. 2016;26:371–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mann DM, Yates PO, Marcyniuk B. A comparison of nerve cell loss in cortical and subcortical structures in Alzheimer’s disease. J Neurol Neurosurg Psychiatry. 1986;49:310–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Curcio CA and Kemper T. Nucleus raphe dorsalis in dementia of the Alzheimer type: neurofibrillary changes and neuronal packing density. J Neuropathol Exp Neurol. 1984;43:359–368. [DOI] [PubMed] [Google Scholar]
- 23.Ishii T. Distribution of Alzheimer’s neurofibrillary changes in the brain stem and hypothalamus of senile dementia. Acta Neuropathol. 1966;6:181–187. [DOI] [PubMed] [Google Scholar]
- 24.Halliday GM, McCann HL, Pamphlett R, et al. Brain stem serotonin-synthesizing neurons in Alzheimer’s disease: a clinicopathological correlation.Acta Neuropathol. 1992;84:638–650. [DOI] [PubMed] [Google Scholar]
- 25.D’Amato RJ, Zweig RM, Whitehouse PJ, et al. Aminergic systems in Alzheimer’s disease and Parkinson’s disease.Ann Neurol. 1987;22:229–236. [DOI] [PubMed] [Google Scholar]
- 26.Durisko Z, Mulsant BH, Andrews PW.An adaptationist perspective on the etiology of depression.J Affect Disord.2015;172:315–323. [DOI] [PubMed] [Google Scholar]
- 27.Kaufman J, Sullivan GM, Yang J, et al. Quantification of the serotonin 1A receptor using PET: identification of a potential biomarker of major depression in males. Neuropsychopharmacology. 2015;40:1692–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gray NA, Milak MS, Delorenzo C, et al. Antidepressant treatment reduces serotonin-1A autoreceptor binding in major depressive disorder. Biol Psychiatry. 2013;74:26–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sullivan GM, Oquendo MA, Milak M, et al. Positron emission tomography quantification of serotonin1a receptor binding in suicide attempters with major depressive disorder. JAMA Psychiatry. 2015;72:169–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gaudin E, Toussaint M, Thibaudeau C, Paille M, Fontaine R, Lecomte R. Performance simulation of an ultrahigh resolution brain PET scanner using 1.2-mm pixel detectors. IEEE Transactions on Radiation and Plasma Medical Sciences.2018;3:334–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rullmann M, Dukart J, Hoffmann KT, et al. Partial-volume effect correction improves quantitative analysis of 18F-florbetaben β-amyloid PET scans. J Nucl Med. 2016;57:198–203. [DOI] [PubMed] [Google Scholar]
- 32.Cherry SR, Jones T, Karp JS, Qi J, Moses WW, Badawi RD. Total-body PET:maximizing sensitivity to create new opportunities for clinical research and patient care.J Nucl Med.2018;59:3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karp JS, Surti S, Daube-Witherspoon ME, et al. Performance of a brain PET camera based on Anger-logic gadolinium oxyorthosilicate detectors. J Nucl Med. 2003;44:1340–1349. [PubMed] [Google Scholar]
- 34.Morimoto Y, Ueno Y, Takeuchi W, et al. Development of a 3D brain PET scanner using CdTe semiconductor detectors and its first clinical application. IEEE Trans Nucl Sci. 2011;58:2181–2189. [Google Scholar]
- 35.Tashima H, Ito H, Yamaya T. A proposed helmet-PET with a jaw detector enabling high-sensitivity brain imaging. In: IEEE Nuclear Science Symposium and Medical Imaging Conference. IEEE; 2013:9–11. [Google Scholar]
- 36.Tashima H, Yoshida E, Nishikido F, et al. Development of the Helmet - Chin PET Prototype. In: IEEE Nuclear Science Symposium and Medical Imaging Conference. IEEE; 2015:1–3. [Google Scholar]
- 37.Watanabe M, Saito A, Isobe T, et al. Performance evaluation of a high-resolution brain PET scanner using four-layer MPPC DOI detectors. Phys Med Biol. 2017;62:7148–7166. [DOI] [PubMed] [Google Scholar]
- 38.Yamaya T, Hagiwara N, Obi T, et al. Preliminary resolution performance of the prototype system for a 4-Layer DOI-PET scanner: JPET-D4. IEEE Trans Nucl Sci. 2006;53:1123–1128. [Google Scholar]
- 39.Sarasola I, Rato Mendes P, García De Acilu P, et al. PET demonstrator for a human brain scanner based on monolithic detector blocks. IEEE Trans Nucl Sci. 2011;58:2190–2197. [Google Scholar]
- 40.Mikhaylova E, De Lorenzo G, Chmeissani M, et al. Simulation of the expected performance of a seamless scanner for brain pet based on highly pixelated CdTe detectors. IEEE Trans Med Imaging. 2014;33:332–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dillon KA, Gross-Isseroff R, Israeli M, Biegon A. Autoradiographic analysis of serotonin 5-HT1A receptor binding in the human brain postmortem: effects of age and alcohol. Brain Res. 1991;554:56–64. [DOI] [PubMed] [Google Scholar]
- 42.Beliveau V, Ganz M, Feng L, et al. A high-resolution in vivo atlas of the human brain’s serotonin system. J. Neurosci 2017;37:120–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Savli M, et al. Normative database of the serotonergic system in healthy subjects using multi-tracer PET. Neuroimage. 2012;6(1):447–459. [DOI] [PubMed] [Google Scholar]
- 44.Moses WW. Fundamental limits of spatial resolution in PET. Nucl Instrum Methods Phys Res A. 2011;648:S236–S240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Van Sluis J, De Jong J, Schaar J, et al. Performance characteristics of the digital Biograph vision PET/CT system. J Nucl Med. 2019;60:1031–1036. [DOI] [PubMed] [Google Scholar]
- 46.Drevets WC, Frank E, Price JC, et al. PET imaging of serotonin 1A receptor binding in depression. Biol Psychiatry. 1999;46:1375–1387. [DOI] [PubMed] [Google Scholar]
- 47.Greve DN, Salat DH, Bowen SL, et al. Different partial volume correction methods lead to different conclusions: an 18F-FDG-PET study of aging. NeuroImage. 2016;132:334–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cysouw MC, Kramer GM, Schoonmade LJ, Boellaard R, de Vet HC, Hoekstra OS. Impact of partial-volume correction in oncological PET studies: a systematic review and meta-analysis. EJNMMI. 2017;44:2105–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schwarz CG, Gunter JL, Lowe VJ, et al. A comparison of partial volume correction techniques for measuring change in serial amyloid pET SUVR. J Alzheimer’s Dis. 2019;67:181–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alavi A, Werner TJ, Høilund-Carlsen PF, Zaidi H. Correction for partial volume effect is a must, not a luxury, to fully exploit the potential of quantitative pet imaging in clinical oncology. Mol Imaging Biol. 2018;20:1–3. [DOI] [PubMed] [Google Scholar]
- 51.Nieuwenhuis S, Aston-Jones G, Cohen JD.Decision making, the P3, and the locus coeruleus–norepinephrine system. Psychol Bull. 2005;131(4):510–532. [DOI] [PubMed] [Google Scholar]
- 52.Aston-Jones G, Cohen JD. An integrative theory of locus coeruleus-norepinephrine function: adaptive gain and optimal performance. Annu Rev Neurosci. 2005;28:403–450. [DOI] [PubMed] [Google Scholar]
- 53.Redmond DE, Huang YH. II. New evidence for a locus coeruleus-norepinephrine connection with anxiety. Life Sci. 1979;25:2149–2162. [DOI] [PubMed] [Google Scholar]
- 54.Pietrzak RH, Gallezot JD, Ding YS, et al. Association of post-traumatic stress disorder with reduced in vivo norepinephrine transporter availability in the locus coeruleus. JAMA Psychiatry. 2013;70:1199–1205. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 55.Satoh A, Iijima KM.Roles of tau pathology in the locus coeruleus (LC) in age-associated pathophysiology and Alzheimer’s disease pathogenesis:potential strategies to protect the LC against aging. Brain Res. 2019;1702:17–28. [DOI] [PubMed] [Google Scholar]
- 56.Giorgi FS, Saccaro LF, Galgani A, et al. The role of locus coeruleus in neuroinflammation occurring in Alzheimer’s disease. Brain Res Bull. 2019;153:47–58. [DOI] [PubMed] [Google Scholar]
- 57.Betts MJ, et al. Locus coeruleus imaging as a biomarker for noradrenergic dysfunction in neurodegenerative diseases. Brain. 2019;142:2558–2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.German DC, Manaye KF, White CL III, et al. Disease-specific patterns of locus coeruleus cell loss. Ann Neurol. 1992;32:667–676. [DOI] [PubMed] [Google Scholar]
- 59.Grudzien A, Shaw P, Weintraub S, Bigio E, Mash DC, Mesulam MM.Locus coeruleus neurofibrillary degeneration in aging, mild cognitive impairment and early Alzheimer’s disease. Neurobiol Aging. 2007;28:327–335. [DOI] [PubMed] [Google Scholar]
- 60.Tona KD, van Osch MJ, Nieuwenhuis S, Keuken MC.Quantifying the contrast of the human locus coeruleus in vivo at 7 tesla MRI. PLoS One. 2019;14:10–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Logan J, Wang G-j, Telang F, et al. Imaging the norepinephrine transporter in humans with (S,S)-[11C]O-methyl reboxetine and PET: problems and progress. Nucl Med Biol. 2007;34:667–679. [DOI] [PubMed] [Google Scholar]
- 62.Liebe T, Kaufmann J, Li M, Skalej M, Wagner G, Walter M. In vivo anatomical mapping of human locus coeruleus functional connectivity at 3 T MRI. Hum Brain Mapp. 2020;41:2136–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Keren NI, Lozar CT, Harris KC, Morgan PS, Eckert MA. In vivo mapping of the human locus coeruleus. NeuroImage. 2009;47:1261–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mohammadi I, Castro IF, Correia PM, Silva AL, Veloso JF. Minimization of parallax error in positron emission tomography using depth of interaction capable detectors: Methods and apparatus. Biomed Phys Eng Express. 2019;5:062001. [Google Scholar]
- 65.LaBella A, Cao X, Petersen E, et al. High-resolution depth-encoding PET detectormodule with prismatoid light-guide array. J Nucl Med. 2020;61:1528–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.LaBella A, Zhao W, Lubinsky R, Goldan AH. Prismatoid light guide array for enhanced gamma ray localization in PET: a Monte Carlo simulation study. Phys Med Biol. 2020;65:18LT01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.LaBella A, Tavernier S, Woody C, Purschke M, Zhao W, Goldan AH. Towards 100 ps coincidence time resolution using multiple timestamps in depth-encoding PET modules: a Monte Carlo simulation study. IEEE Trans Rad Plasma Med Sci. 2021;5(5):679–686. [Google Scholar]
- 68.Cao X, La Bella A, Zeng X, Zhao W, Goldan AH. Depth of interaction and coincidence time resolution characterization of ultrahigh resolution time-of-flight prism-PET modules. IEEE Trans Radiat Plasma Med Sci. 2022;6(5):529–536. [Google Scholar]
- 69.LaBella A, Cao X, Zeng X, Zhao W, Goldan AH.Sub-2 mm depth of interaction localization in PET detectors with prismatoid light guide arrays and single-ended readout using convolutional neural networks. Med Phys. 2021;48(3):1019–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Petersen E, LaBella A, Howansky A, Cao X, Zhao W, Goldan A. Compton scatter decomposition and recovery in a Prism-PET module. J Nucl Med. 2020;61(1):1510. [Google Scholar]
- 71.Wang Z, Cao X, Zeng X, et al. A high resolution and high sensitivity Prism-PET brain scanner with non-cylindrical decagon geometry. J Nucl Med. 2021;62(1):1136. [Google Scholar]
- 72.LaBella A, Zhao W, Lubinsky R, Goldan AH. Prism mirror light guide for enhanced gamma ray localization in PET. In: 2019 IEEE Nuclear Science Symposium and Medical Imaging Conference (NSS/MIC). IEEE; 2019:1–4. [Google Scholar]
- 73.Varnäs K, Halldin C, Hall H. Autoradiographic distribution of serotonin transporters and receptor subtypes in human brain. Hum Brain Mapp. 2004;22:246–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hall H, Lundkvist C, Halldin C, et al. Autoradiographic localization of 5-HT1A receptors in the post-mortem human brain using [3H]WAY-100635 and [11C]WAY-100635. Brain Res. 1997;745:96–108. [DOI] [PubMed] [Google Scholar]
- 75.Schou M, Halldin C, Pike VW, et al. Post-mortem human brain autoradiography of the norepinephrine transporter using (S,S)-[18F]FMeNER-D2. Eur Neuropsychopharm. 2005;15:517–520. [DOI] [PubMed] [Google Scholar]
- 76.Jan S, Benoit D, Becheva E, et al. GATE: a simulation toolkit for PET and SPECT. Phys Med Biol. 2004;49:4543–4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Badawi RD, Shi H, Hu P, et al. First human imaging studies with the EXPLORER total-body PET scanner. J Nucl Med. 2019;60:299–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vandenberghe S, Moskal P, Karp JS. State of the art in total body PET. EJNMMI Physics. 2020;7:1–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zein SA, Karakatsanis NA, Conti M, Nehmeh SA. Monte Carlo simulation of the siemens biograph vision PET with extended axial field of view using sparse detector module rings configuration. IEEE Trans Radiat Plasma Med Sci. 2020;5:331–342. [Google Scholar]
- 80.NEMA N. NU 2-2012: Performance Measurements of Positron Emission Tomographs. Rosslyn, VA: National Electrical Manufacturers Association; 2012. [Google Scholar]
- 81.Zubal IG, Harrell CR, Smith EO, Rattner Z, Gindi G, Hoffer PB. Computerized three-dimensional segmented human anatomy. Med Phys. 1994;21:299–302. [DOI] [PubMed] [Google Scholar]
- 82.Thielemans K, Tsoumpas C, Mustafovic S, et al. STIR: software for tomographic image reconstruction release 2.Phys Med Biol. 2012;57(4):867. [DOI] [PubMed] [Google Scholar]
- 83.Yamaya T, Hagiwara N, Obi T, et al. DOI-PET image reconstruction with accurate system modeling that reduces redundancy of the imaging system. IEEE Trans Nucl Sci. 2003;50:1404–1409. [Google Scholar]
- 84.Merlin T, Stute S, Benoit D, et al. CASToR: a generic data organization and processing code framework for multi-modal and multi-dimensional tomographic reconstruction. Phys Med Biol. 2018;63:185005. [DOI] [PubMed] [Google Scholar]
- 85.Fernandes P, Regala J, Correia F, Gonçalves-Ferreira AJ. The human locus coeruleus 3-D stereotactic anatomy. Surg Radiol Anat. 2012;34:879–885. [DOI] [PubMed] [Google Scholar]
- 86.Hornung J-P.The human raphe nuclei and the serotonergic system. J Chem Neuroanat. 2003;26:331–43. [DOI] [PubMed] [Google Scholar]
- 87.Murphy PR, O’Connell RG, O’Sullivan M, Robertson IH, Balsters JH. Pupil diameter covaries with BOLD activity in human locus coeruleus. Hum Brain Mapp. 2014;35:4140–4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Anand A, Jones SE, Lowe M, Karne H, Koirala P. Resting state functional connectivity of dorsal raphe nucleus and ventral tegmental area in medication-free young adults with major depression. Front Psychiatry. 2019;259:765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Morris LS, Tan A, Smith DA, et al. Sub-millimeter variation in human locus coeruleus is associated with dimensional measures of psychopathology:An in vivo ultra-high field 7-Tesla MRI study. NeuroImage Clin. 2020;25:102148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Carson RE, Wu Y, Lang L, et al. Brain uptake of the acid metabolites of F-18-Labeled way 100635 analogs. J Cereb Blood Flow Metab. 2003;23:249–260. [DOI] [PubMed] [Google Scholar]
- 91.Mukherjee J, Bajwa AK, Wooten DW, et al. Comparative assessment of 18F-Mefway as a serotonin 5-HT1A receptor PET imaging agent across species: rodents, nonhuman primates, and humans. J Comp Neurol. 2016;524:1457–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Friston KJ. Statistical parametric mapping. Neuroscience databases. Boston, MA: Springer; 2003:237–250. [Google Scholar]
- 93.MacDonald LR, Dahlbom M. Parallax correction in PET using depth of interaction information. IEEE Trans Nucl Sci. 1998;45:2232–2237. [Google Scholar]
- 94.Kim K, Dutta J, Groll A, El Fakhri G, Meng LJ, Li Q. A novel depth-of-interaction rebinning strategy for ultrahigh resolution PET. Phys Med Biol. 2018;63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang M, Zhou J, Yang Y, Rodríguez-Villafuerte M, Qi J.Efficient system modeling for a small animal PET scanner with tapered DOI detectors. Phys Med Biol. 2015;61:461–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gillam JE, Solevi P, Oliver JF, et al. Sensitivity recovery for the AX-PET prototype using inter-crystal scattering events. Phys Med Biol. 2014;59:4065. [DOI] [PubMed] [Google Scholar]
- 97.Levin CS. Promising new photon detection concepts for high-resolution clinical and preclinical PET.J Nucl Med. 2012;53:167–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Levin CS. New imaging technologies to enhance the molecular sensitivity of positron emission tomography. Proc IEEE. 2008;96:439–467. [Google Scholar]
- 99.Petersen EW, LaBella A, Howansky A, Zhao W, Goldan AH. Compton decomposition and recovery in a Prism-PET Detector Module. In: 2020 IEEE Nuclear Science Symposium and Medical Imaging Conference (NSS/MIC). IEEE; 2020:1–4. [Google Scholar]
- 100.Bettinardi V, Castiglioni I, De Bernardi E, Gilardi MC.PET quantification: Strategies for partial volume correction. Clin Transl Imaging. 2014;2:199–218. [Google Scholar]
- 101.Wienhard K, et al. The ECAT HRRT: performance and first clinical application of the new high resolution research tomograph. IEEE Trans Nucl Sci. 2002;49:104–110. [Google Scholar]
- 102.Catana C. Development of dedicated brain PET imaging devices: recent advances and future perspectives. J Nucl Med. 2019;60:1044–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ding YS, et al. PET imaging of the effects of age and cocaine on the norepinephrine transporter in the human brain using (S, S)-[11C] O-methylreboxetine and HRRT. Synapse. 2010;64:30–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhang X, Zhou J, Cherry SR, Badawi RD, Qi J. Quantitative image reconstruction for total-body PET imaging using the 2-meter long EXPLORER scanner. Phys Med Biol. 2017;62:2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Schmall JP, Karp JS, Werner M, Surti S. Parallax error in longaxial field-of-view PET scanners—a simulation study.Phys Med Biol. 2016;61:5443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Berg E, Roncali E, Kapusta M, Du J, Cherry SR. A combined time-of-flight and depth-of-interaction detector for total-body positron emission tomography. Med Phys. 2016;43:939–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Arendt T, Brückner MK, Morawski M, Jäger C, Gertz HJ. Early neurone loss in Alzheimer’s disease: cortical or subcortical? Acta Neuropathol Commun. 2015;3:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Braak H, Del Tredici K.Where, when, in what form does sporadic Alzheimer’s disease begin?.Curr Opin Neurol.2012;25:708–714. [DOI] [PubMed] [Google Scholar]
- 109.Betts MJ, et al. Locus coeruleus imaging as a biomarker for noradrenergic dysfunction in neurodegenerative diseases. Brain. 2019;142:2558–2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ehrenberg AJ, Nguy AK, Theofilas P, et al. Quantifying the accretion of hyperphosphorylated tau in the locus coeruleus and dorsal raphe nucleus: the pathological building blocks of early Alzheimer’s disease. Neuropath Appl Neuro. 2017;43:393–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Heneka MT, Ramanathan M, Jacobs AH, et al. Locus ceruleus degeneration promotes Alzheimer pathogenesis in amyloid precursor protein 23 transgenic mice. J Neurosci. 2006;26:1343–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Liu KY, et al. FDG-PET assessment of the locus coeruleus in Alzheimer’s disease. Neuroimage: Reports. 2021;1:100002. [DOI] [PMC free article] [PubMed] [Google Scholar]


