Figure 4. Overview of tools to characterize diverse types of genomic loci.
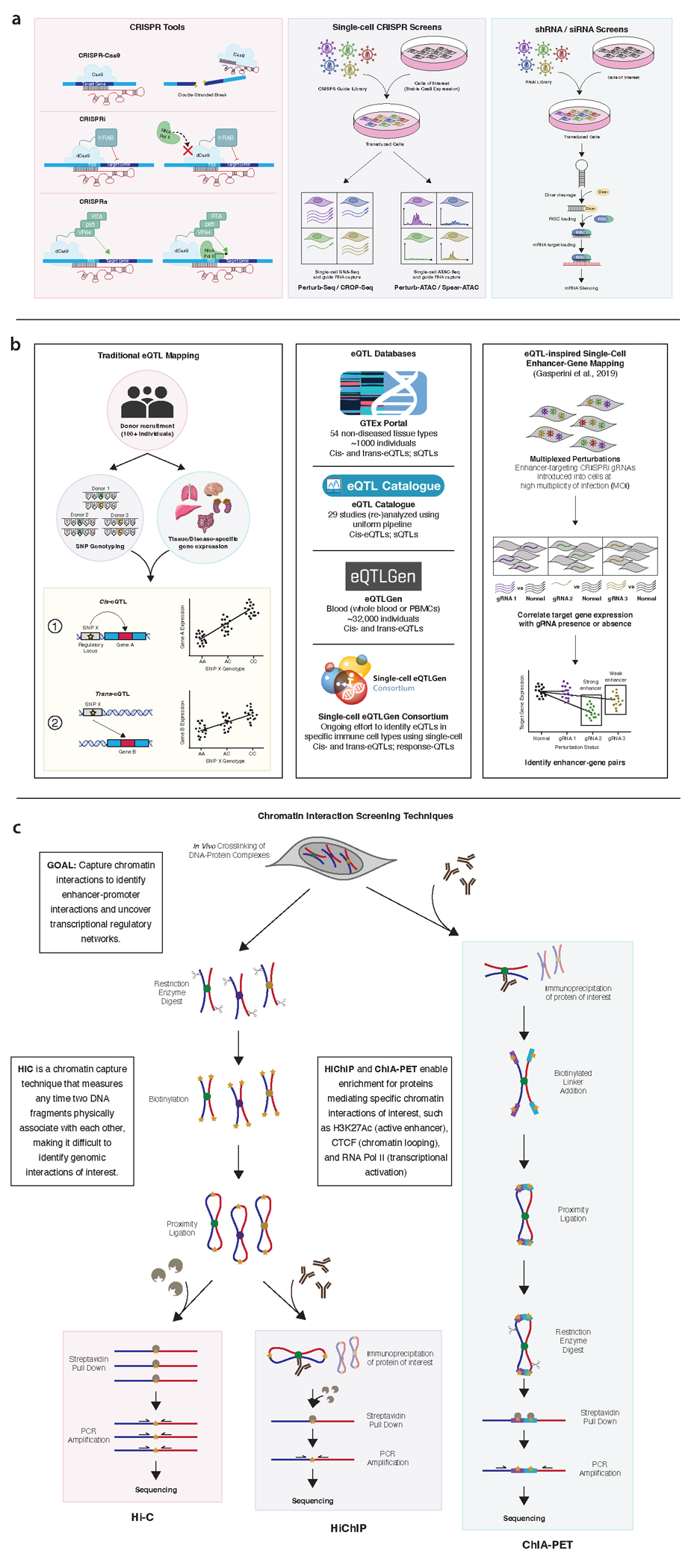
(a) CRISPR tools, including CRISPRi and CRISPRa, have been used to characterize coding and noncoding regions. CRISPR screens like Perturb-Seq can be leveraged to study the effect of a CRISPR perturbation on gene expression in a high-throughput, single-cell manner. shRNA and siRNA screens can similarly be used to test for the effect of gene silencing, this time at the RNA level, on a phenotype. (b) Colocalization of GWAS susceptibility loci with eQTLs is a common strategy for identifying target genes for non-coding variants/loci. Traditional eQTL mapping uses hundreds of individuals to identify associations between a variant and a change in gene expression. eQTL databases include eQTLGen and GTEx. Single-cell eQTL mapping using high MOI CRISPR perturbation can bypass the need for hundreds of human samples. (c) Hi-C, HiChIP, and ChIA-PET are chromatin capture techniques used to identify physical contacts between DNA to predict enhancer-promoter interactions. HiChIP and ChIA-PET allow for capture of specific proteins to enrich for enhancer-promoter loops.
