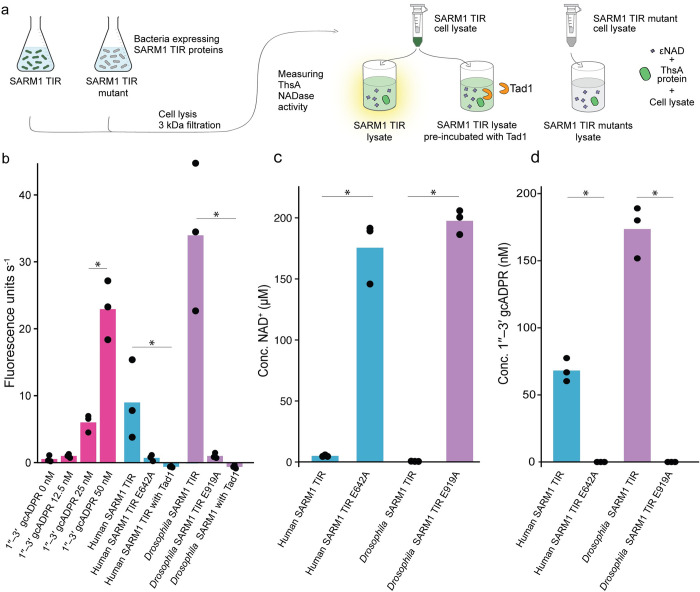
Fig 1. Detection of 1′′–3′ gcADPR in lysates of bacteria expressing the SARM1 TIR domain.
(a) Schematic representation of the experiment. Filtered lysates of cells expressing SARM1 TIR or SARM1 TIR active site mutants were tested for activation of the ThsA protein. NADase activity of ThsA was measured using a nicotinamide 1,N6-ethenoadenine dinucleotide (εNAD) cleavage fluorescence assay. (b) Activity of purified ThsA protein from B. cereus, incubated with increasing concentration of 1′′–3′ gcADPR as well as with lysates derived from bacteria that express the SARM1 TIR domain from human and D. melanogaster. Data are also shown for lysates from bacteria expressing the SARM1 TIR domains with catalytic site mutations, as well as lysates that were pre-incubated with Tad1. Bars represent the mean of three independent replicates, with individual data points overlaid. Asterisks indicate a statistically significant difference (one-way ANOVA followed by pairwise multiple comparison analysis according to Tukey’s honest significant difference criterion, P < 0.05). (c) LC-MS analysis showing concentrations of NAD+ in cell lysates extracted from E. coli expressing human and Drosophila SARM1 TIR domains. Control cells in this experiment express SARM1 TIR domains with catalytic site mutations. Bar graphs represent the average of three independent replicates, with individual data points overlaid. Asterisk marks statistically significant increase (Student’s t-test, two-sided, P <0.05). (d) LC-MS analysis showing concentrations of 1′′–3′ gcADPR in cell lysates extracted from E. coli expressing human and Drosophila SARM1 TIR domains. The cells used in this experiment are as in panel c. Bar graphs represent the average of three independent replicates, with individual data points overlaid. Asterisk marks statistically significant decrease (Student’s t-test, two-sided, P <0.05).