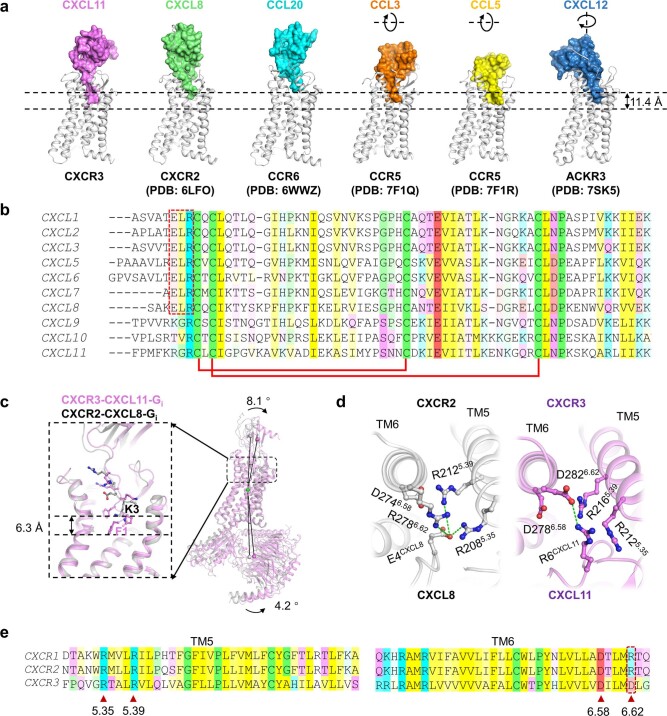
Extended Data Fig. 7. Structural basis for the chemokine selectivity of CXCR2 and CXCR3.
a Comparation of the insertion depth of CXCL11 (violet), CXCL8 (green), CCL20 (cyan), CCL3 (orange), CCL5 (yellow), and CXCL12 (blue). The receptors are shown as cartoon and colored gray, while the chemokines are shown in surface model. The pseudo rotation axis and orientation for CCL3, CCL5, and CXCL12 compared to CXCL11 are shown. b Sequence alignment of CXCL1-3 and 5-11. The disulfide bonds are indicated as red line links. The ELR motif in CXCL1-3 and 5-8 are indicated by a dotted box. c Comparation of CXCR3-CXCL11-Gi (violet) and CXCR2-CXCL8-Gi (gray). The angles are measured using the mass centers of the receptors, the chemokines, and the Gi complexes as reference points. d Residues in CXCR2 and CXCR3 that are involved in the binding of E4CXCL8 and R6CXCL11. The structure of CXCR2 and CXCR3 are shown as cartoon and colored gray and violet, respectively. Residues involved in interactions are shown as sticks, and interactions are indicated by green dashes. e Sequence alignment of TM5 and TM6 in CXCR1, CXCR2 and CXCR3. Key residues in TM5 and TM6 are indicated by red-colored triangles. In b and e, positively charged residues, negatively charged residues, non-charged hydrophilic residues, and hydrophobic residues are colored cyan, red, violet, and yellow, respectively. Residues cystine, proline, and glycine are colored green. The value of conservation visibility is set to 30% in JalView.