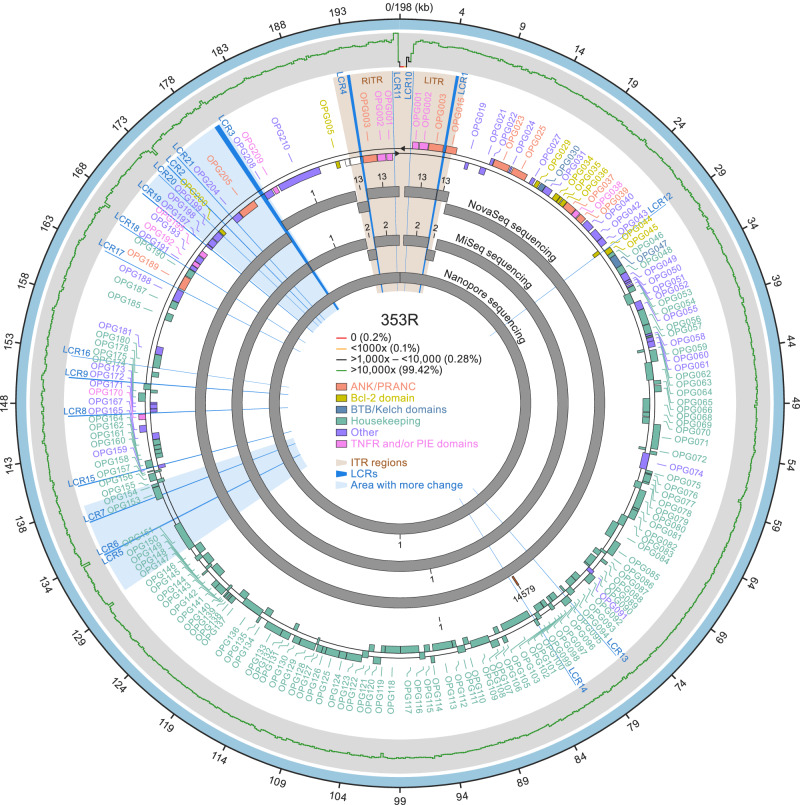
Fig. 1. De novo assembly of subclade IIb lineage B.1 monkeypox virus (MPXV) genome sequence 353R.
A visual representation of the fully annotated MPXV isolate 353R genome (based on the subclade IIb lineage A reference isolate MPXV-M5312_HM12_Rivers genome sequence annotation). Shown are (from the outside to the inside): high-quality genome (HQG) hybrid assembly (wide outer light blue ring); sequencing coverage distribution graph (thin ragged line [green: ≥10,000x, 99.42%; black: 1000x–10,000, 0.28%; orange: <1000x–10, 0.1%; red: <10–0, 0.2%]); orthologous poxvirus gene (OPG) annotations according to the standardized nomenclature32 (lettering and shaded boxes [orange: ANK/PRANC (N-terminal ankyrin protein with PRANC domain) inverted terminal repetition [ITR] regions; gold: Bcl-2 domain; blue: BTB/Kelch domains; green: housekeeping; purple: other; pink: TNFR and/or PIE domains); contigs from NovaSeq, MiSeq, and nanopore sequencing (wide inner gray rings). Additionally, radial lines and shading that originate in the center and reach outward on the white background indicate low-complexity regions (LCRs; royal blue) and areas with more change (light blue).