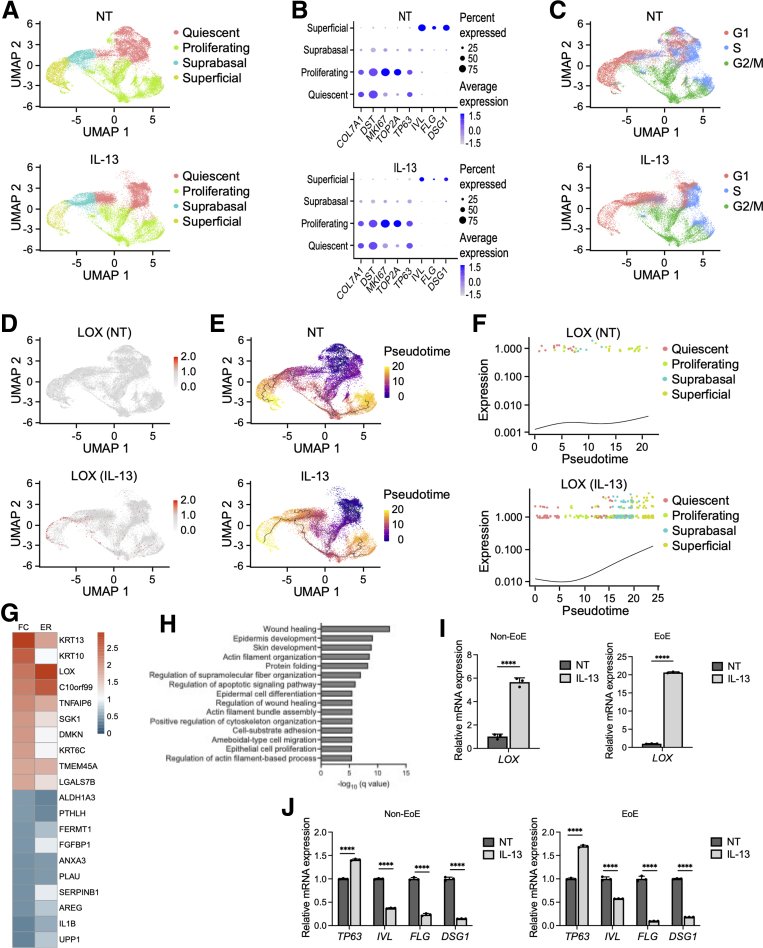
Figure 1.
scRNA-seq from PDOs reveals specific distribution of LOX in esophageal epithelium. (A–H) scRNA-seq analyses based on PDOs from 3 non-EoE patients. PDOs were cultured with or without IL13 (10 μg/mL) from days 7 to 11 and then harvested on day 11. (A) UMAP plots displaying 4 distinct cell types (quiescent basal, proliferating basal, suprabasal, and superficial) in nontreated (NT) and IL13-stimulated PDOs. (B) Dot plots showing the expression of COL7A1, DST, MKI67, TOP2A, TP63, IVL, FLG, and DSG1 in NT and IL13-stimulated PDOs. (C) UMAP plots showing cell-cycle phases in NT and IL13-stimulated PDOs. (D) UMAP plots showing the expression of LOX in NT and IL13-stimulated PDOs. (E) Inferred cell fate trajectories in NT and IL13-stimulated PDOs. Cells are colored by pseudotime value. Dark purple and bright yellow imply the earliest cells and the latest cells, respectively. (F) Expression kinetics for LOX across the pseudotime axis in NT and IL13-stimulated PDOs. (G) Heatmap representation of fold change (FC) and expression ratio (ER) of the average expression across all cells of the top 10 differentially expressed genes in LOX-expressing cells (LOX+) compared with LOX nonexpressing (LOX-) cells in the superficial cluster of IL13-stimulated PDOs. (H) Gene Ontology analysis of DEG profiles of LOX+ cells compared with LOX- cells in the superficial cluster of IL13-stimulated PDOs. (I) Relative expression, via qRT-PCR, of LOX gene in EoE or non-EoE PDOs, cultured with or without IL13 (representative results are shown, total n = 2/2 EoE/non-EoE lines). (J) Relative expression, via qRT-PCR, of TP63, IVL, FLG, and DSG1 genes in EoE or non-EoE PDOs, cultured with or without IL13 (representative results are shown, total n = 2/1 EoE/non-EoE lines).. ∗∗∗∗P < .0001