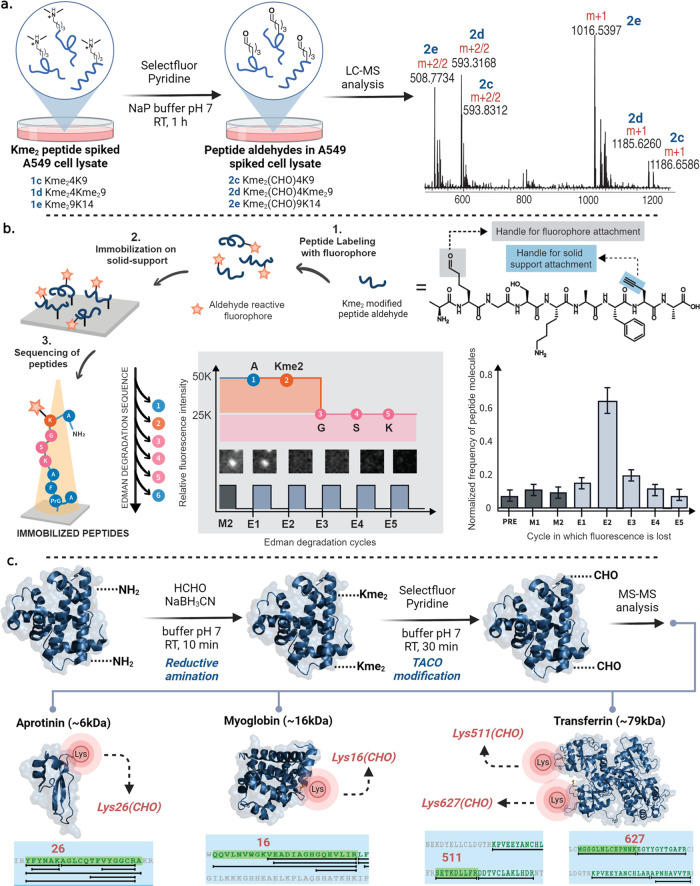
Figure 5.
Labeling and identification of Kme2 peptides and proteins in a complex mixture. (a) Selective modification of Kme2 peptides in a complex mixture of A549 lung cancer cell lysate to peptide aldehydes in a quantitative manner. Reaction conditions: To 100 μg of A549 whole cell lysate in 200 μL of NaP buffer pH 7.0 was added 0.1 mg of each histone peptide followed by addition of pyridine (30 equiv with respect to peptides) and selectfluor (10 equiv with respect to peptides). The reaction mixture was stirred for 1 h. (b) Application of TACO in the identification of Kme2 sites on a peptide by single molecule fluorosequencing. The left panel provides an exemplary view of the single fluorescent peptide molecule through cycles of Edman degradation. The right panel indicates the normalized counts of the labeled Kme2 peptide where its fluorescence intensity was lost. (c) Selective modification of chemically generated Kme2 proteins by TACO chemistry followed by the identification of Kme2 sites on proteins by LC-MS/MS analysis.