Fig. 1.
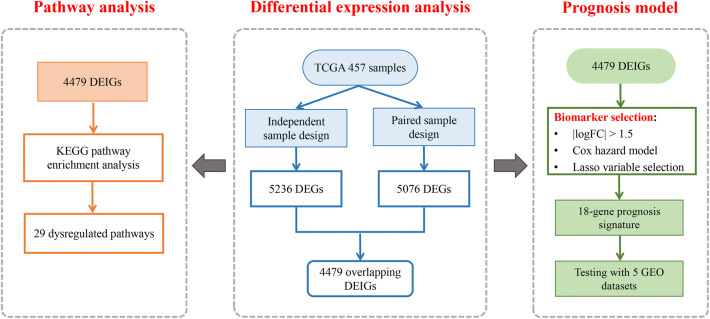
A flowchart of the study design. First, we developed a novel analysis strategy by combining two different study designs to detect differential expression of immune genes between tumor and normal tissues in TCGA LUAD patients. Next, the significant DEGs were used to perform a pathway enrichment analysis. Further, we constructed a prognosis model with the identified DEGs aiming to distinguish high-risk populations and guide early treatment. The predictive model was validated in five independent lung cancer gene expression datasets