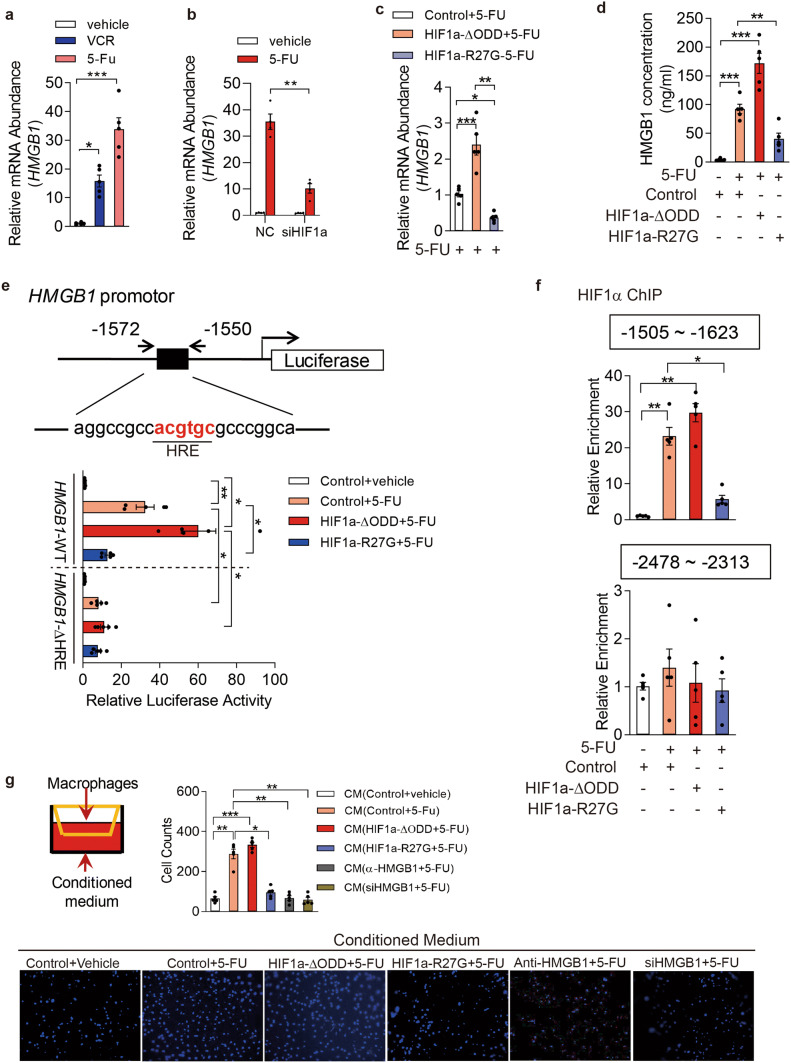
Fig. 2.
HIF1α drives HMGB1 expression in 5-FU-treated gastric cancer cells to exacerbate macrophage infiltration. a qRT-PCR analysis of HMGB1 mRNA levels of AGS cells in the presence of 5-FU (0.1 mg/ml), VCR (0.1 nmol/l) or vehicle control for 24 h. b qRT-PCR analysis of HMGB1 mRNA levels of AGSs in the presence of 5-FU or vehicle. The AGS cells were transfected with siRNA targeted at HIF1a (siHIF1a) or negative control (NC) prior to 5-FU or vehicle treatment. c, d, AGS cells were transfected with empty vector (control) or constructs expressing human HIF1a mutants HIF1a-ΔODD or HIF1a-R27G. (c) qRT-PCR analysis of HMGB1 mRNA levels of 5-FU-treated cells and d ELISA determined the HMGB1 protein levels in the cell culture of AGS cells after treated by vehicle or 5-FU. e AGS cells were transfected with reporter constructs within which luciferase expression is driven under HMGB1 full-length promoter (2000 bp upstream of TSS) or HRE-truncated promoter in combination with empty vector (control) or constructs expressing HIF1a-ΔODD or HIF1a-R27G, respectively. Luciferase activity is presented as fold change after normalizing Luc values to Renilla activity (RLU). The values of vehicle control group were set as “1”. f ChIP assays were conducted by the using of IgG or anti-HIF1α antibody in cell lysis from AGS cells which were transfected with control vector or constructs expressing HIF1a-ΔODD or HIF1a-R27G followed with treatments of 0.1 mg/ml 5-FU or vehicle. qRT-PCR analyses of immunoprecipitated DNA were performed using the primers which were designed to amplify the indicated region of the HMGB1 promoter. g Representative images of transmembraned macrophages counterstained with DAPI. Transwell assays of macrophages were conducted in the presence of conditioned media derived from the cell culture of AGS cells which were previously subjected to indicated treatments. All data are shown as mean ± SEM. *p < 0.05; **p < 0.01; or ***p < 0.001 by unpaired two-tailed Student’s t test or one-way ANOVA