Abstract
Tumor-associated macrophages (TAMs) are important regulators of the complex interplay between immune system and breast cancer. TAMs fuel the cancer progression and metastasis by reprogramming their specific functional phenotype in cancer settings. Therefore, it is important to clarify the mechanisms of shaping specific functional phenotype of macrophages in tumor milieu. LncRNA profiles of TAMs were identified by LncRNA microarray. Flow cytometry was used to detect the surface markers of TAMs. The co-localization among lincRNA-p21, p53 and Mouse Double Minute 2 (MDM2) was identified by FISH probe and immunofluorescence. PyVT-MMTV and BALB/c mice were used for in vivo analysis. In the present work, we found that lincRNA-p21 significantly up-regulated in 4T1 educated macrophages. LincRNA-p21 knockdown facilitated macrophage polarization into pro-inflammatory M1 in tumor microenvironment, which might be caused by MDM2 eliciting proteasome-dependent degradation to p53 and activated NF-κB and STAT3 pathway. TAMs with lincRNA-p21 knockdown induced cancer cell apoptosis, inhibited tumor cell migration and invasion. In vivo, lincRNA-p21 knockdown macrophage adoptive transfer could alleviate breast cancer progression. Our results indicated that lincRNA-p21 was a key regulator of TAMs function in tumor milieu. Our data also shed a light on novel therapeutic targets of tumors characterized by monocytes/macrophages infiltration.
Keywords: TAMs, lincRNA-p21, Breast cancer, p53, Tumor microenvironment
Introduction
Breast cancer is one of the most common malignant cancer in women [1]. Recently, the incidence of breast cancer has been increasing and become another killer of women [2]. Invasive breast cancer is more dangerous because of easy metastasis and recurrence [3]. Although it can be treated by surgery or chemotherapy, the chances of recovery decline exponentially as the disease progresses [4]. Currently, many treatments only focus on breast cancer cells and neglect to improve the tumor microenvironment. A lot of data have shown that tumor microenvironment plays important roles in breast cancer development [5].
Tumor microenvironment is a complex environment for tumor cells to survive, which is mainly composed of tumor cells, immune cells, stromal cells and extracellular matrix [6]. Macrophages are an important population in tumor stroma [7]. Generally, they are from mononuclear or tissue-resident macrophages and display specific phenotypic characteristics [8]. Traditionally, macrophages are considered as the first protective barrier and keeping homeostasis, they also can directly kill tumor cells, induce or regulate immune response [9]. Recent data have shown that macrophages are extraordinary disparate population that incessantly alter their functional phenotype in response different environment stress [10]. They undergo the “polarization” course or functional shift, express diverse surface markers, produce different inflammatory mediators following microenvironmental stimuli [11]. In solid tumor microenvironment, peripheral blood monocytes infiltrate into tumor tissue through blood vessels and polarize into tumor-associated macrophages (TAMs) which show various phenotypes in tumor development [12]. Tumorigenesis is an extremely complex and multi-step process. Many studies have shown that macrophages play a special role in tumorigenesis [13]. In the primary stage of tumor development, TAMs exert a pro-inflammatory phenotype, initiate an anti-tumor type-1 inflammatory response, and inhibit the growth of tumor cells by producing TNF-α, reactive oxygen species (ROS) or exerting phagocytosis [14, 15]. Conversely, at the advanced stage of tumor development, the educated macrophages by tumor microenvironment secrete IL-10 and TGF-β and restrain the activation of cytotoxic T lymphocyte (CTL) and natural killer (NK) cells [16]. Additionally, the anti-inflammatory macrophages can also secrete a variety of growth factors and proteolytic enzymes and play major roles in tissue remodeling and tumor progression [17].
The mechanism by which peripheral blood monocytes are recruited into the tumor microenvironment and polarized remains unclear. Functional genomics studies have shown that protein-coding genes account for less than 2% of the genome, while more than 98% of the genome is non-coding RNA, including small RNA represented by small interfering RNA (siRNA), small RNA (miRNA), and long non-coding RNA (lncRNA) [18]. LncRNA is a class of non-coding RNA with a length of more than 200 nucleotides, which are found in eukaryotes [19]. LncRNA is previously thought to lack biological functions. Recently, many data have demonstrated that lncRNA plays an extremely important role in the vital movement of organisms, and gene expression and shutdown, cell cycle and ontogenesis regulation [20] as well as immune cells’ differentiation [21]. As well known, p53 is an important regulator of DNA damage and cell cycle arrest [22]. It has been demonstrated that lincRNA-p21 regulates p53-dependent target genes and involve in atherosclerosis development [23]. Some data also show that lincRNA-p21 is decreased in some tumor cells and inflammatory diseases such as rheumatoid arthritis [24, 25]. Additionally, in hypoxic environment, lincRNA-p21 can promote tumor cells’ survival and proliferation by accelerating anaerobic glycolysis of tumor cells [26]. However, lncRNAs’ role in polarization or functional shift of infiltrated macrophages in tumor milieu remains unclear. Therefore, the present work would focus on the changes of lncRNAs in breast cancer infiltrated macrophages and analyze their roles on functional shift of macrophage and breast cancer development.
Materials and methods
Peritoneal macrophage isolation, TAMs’ induction in vitro and cell lines
BALB/c mice were lavaged with 5 mL cold PBS. After washing, the lavage fluid was collected and centrifuged at 1000 rpm for 5 min. The cells were plated on a 12-well plate in RPMI 1640 supplemented with 2% FBS and 500 IU/mL penicillin–streptomycin. After 3 h of culture, plates were washed three times to remove non-adherent cells. The adherent cells were used in subsequent experiments.
Mouse breast cancer cell line 4T1 and Lewis lung carcinoma (LLC) were purchased from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). Cancer cells were seeded at 6-well plates with 1 mL RPMI 1640 supplemented with 10% FBS and cultured for 48 h; the supernatant was collected to treat macrophages for 48 h. After identification, the treated macrophages showed M2 phenotype and these cells were considered as TAMs. All cells were passaged when cells reached 80%–90% confluence. In all experiments, the cells were seeded at an appropriate density and grew to a confluence of 80%–90% before treatment.
Animal models of tumors
PyVT-MMTV mice were purchased from the Model Animal Research Center of Nanjing University (Nanjing, China) and Specific pathogen-free (SPF) female BALB/c mice (6–8 weeks old) were purchased from Animal Research Center of Jiangsu University. All the mice were bred in Animal Research Center of Jiangsu University (Zhenjiang, China) in compliance with the Guide for the Care and Use of Laboratory Animals” [NIH, 76 FR 91 (May 11, 2011)]. All the protocols were approved by the Committee for Ethical Affairs of Jiangsu University (Zhenjiang, China) and the methods were carried out in "accordance" with the approved guidelines. To establish different tumor models, BALB/c mice were subcutaneously injected in the flank with 4T1 cells (1 × 106/mouse) and macrophages with/without lincRNA-p21 knockdown (1 × 105/mouse) in 200 μL of PBS. Tumor growth was observed with bidirectional tumor measurements using a caliper every 3 days, and tumor volume was calculated using the formula V = ab2/2 (“V” means volume, “a” and “b” represent the length and width, respectively). All the tumor bearing mice were killed, every tumor was isolated and weighed.
Transfection
LincRNA-p21 siRNA was purchased from Shanghai GenePharma Co.,Ltd (Shanghai, China). Cells were transfected using a complex containing 100 nM siRNA and Lipofectamine 2000 (Thermo Fisher Scientific, Waltham, MA, USA). After 6 h transfection, the medium was changed, the fluorescence efficiency was observed using fluorescent microscope.
RNA isolation and RT-qPCR analysis
Total RNA was extracted from cells using TRIzol reagent and reverse transcribed into cDNA according to the manufacturer’s instructions. RT-qPCR was performed on a CFX Connect Real-Time PCR Detection System using iQ SYBR Green Supermix (Bio-Rad, Shanghai, China). Relative gene expression was normalized to endogenous control gene β-actin. The PCR primers were used as following: LincRNA-p21: F 5′- CCTGTCCACTCGCTTTC-3′, R 5′- GGAACTGGAGACGGAATGTC-3′; LincRNA-cox2: F 5′- AAGGAAGCTTGGCGTTGTGA -3′, R 5′- GAGAGGTGAGGAGTCTTATG -3′; LncRNA-A930001c03Rik: F 5′- GAGGCTATCTGAACTCTCTGTGG -3′, R 5′- GAGCATTTTGCTTGCATGATAG -3′; β-actin: F 5′- AGCCATGTACGTAGCCATCC -3′, R 5′- GCTGTGGTGGTGAAGCTGTA -3′.
Western blotting
Cells were collected at indicated points and cell lysates were added and mixed in SDS buffer. Protein extracts were separated in 8%–15% SDS-PAGE and transferred to polyvinylidene fluoride (PVDF) membranes. Membranes were blocked by 5% nonfat dried milk in TBS containing 0.1% Tween 20, and incubated with specific primary antibodies: anti-β-actin (Abcam, Shanghai, China), anti-p53, anti-MDM2, anti-Bax, anti-Bcl-2 and anti-Arginase (Santa Cruz Biotechnology, Shanghai, China), anti-iNOS, anti-Stat3, anti-phospho-Stat3, anti-p65 and anti-phospho-p65 (Cell Signaling Technology, Danvers, MA, USA). Peroxidase-conjugated anti-rabbit IgG or anti-mouse IgG (Abcam, Shanghai, China) was applied to incubate with membranes as the secondary antibody, and the detection were performed using ECL chemiluminescence system (Image Quant LAS4000mini, GE Healthcare, China).
ELISA assay
Mouse peritoneal macrophages with/without lincRNA-p21 knockdown were treated with 4T1 mouse breast tumor cell supernatant (4T1CM) for 48 h, respectively, changed medium and cultured for the other 12 h. The supernatant was collected using for detection of TNF-α, IL-4, IL-6, IL-10 and IL-12 p70 by ELISAs Kit (MultiSciences (Lianke) Biotech Co., Ltd, Hangzhou, China), according to the manufacturer’s instructions.
LncRNA microarray analysis
The lncRNA microarray and the subsequent analysis were completed by Shanghai OE Biotech. Co., Ltd. Briefly, total RNA was extracted from control and 4T1CM treated macrophages, respectively, quantified by NanoDrop ND-2000 (Thermo Scientific, Waltham, MA, USA); the RNA quality was assessed using Agilent Bioanalyzer 2100 (Agilent Technologies Co. Ltd, PaloAlto, CA, USA). The sample labeling and microarray hybridization were performed based on the manufacturer’s instruction.
Feature Extraction software (Version10.7.1.1, Agilent Technologies Co. Ltd, PaloAlto, CA, USA) was used to analyze array images to get raw data. Genespring (Version 13.1, Agilent Technologies Co. Ltd, PaloAlto, CA, USA) was employed to finish the basic analysis with the raw data. The raw data were normalized with the quantile algorithm. Differentially expressed lncRNAs were then identified through fold changes. Gene ontology (GO) and KEGG analysis were applied to determine the roles of these differentially expressed lncRNAs. Finally, hierarchical clustering was performed to show the distinguishable genes' expression pattern.
Flow cytometry analysis
To assess the expression of CD80, CD86, MHC II and CD206 on macrophages with/without lincRNA-p21 knockdown, the cells were treated by 4T1CM. Cells were collected using cold PBS and incubated with anti-mouse CD80-PE, anti-mouse CD86-FITC, anti-mouse MHC II-PE and anti-mouse CD206-APC at 4 °C; all the antibodies were obtained from BD Bioscience (Shanghai, China). After 30 min, cells were re-suspended in PBS and analyzed by flow cytometer (BD Bioscience, Shanghai, China), and the data were analyzed by FlowJo software.
For cellular apoptosis assay, cells were stained with Annexin-V-APC and PI using the Cell Apoptosis Analysis Kit (MultiSciences (Lianke) Biotech Co., Ltd, Hangzhou, China) according to the manufacturer’s instructions. (AnnexinV-APC)−/PI+ cells were considered as necrosis, (AnnexinV-FITC)+/PI- and (AnnexinV-FITC)+/PI+ cells were considered as apoptosis.
Isolation of TAMs from tumor tissues
Tumor tissues were cut into small pieces and incubated for 2 h at 37 °C in 40 mL collagenase constituting of collagenase I, Dnase I and hyaluronidase (Sigma Aldrich, St. Louis, MO, USA), then filtered the tissue and depleted RBC with red blood cell lysis buffer (ACK). Cells were separated using cold PBS and incubated with anti-mouse CD11b-PE and anti-mouse F4/80-FITC (BD Bioscience, USA) and then TAMs isolated by flow cell sorting technique. Macrophages isolated from healthy mice by the macrophage isolating kit (TBD science, Tianjin, China) were used as control.
Cell proliferation, cell migration and wound healing assay
Macrophages with/without lincRNA-p21 knockdown were co-cultured with 4T1 cells in Transwell System for 24 h and 4T1 cell proliferation was detected using the Cell Counting Kit-8(CCK-8) assay Kit (Fcmacs, Nanjing, China), according to the manufacturer's instructions at indicated point.
4T1 cell migration was assessed using 24-well plates with 8 μm pore-size chambers insert (Costar, Cambridge, MA, USA). 5 × 104 4T1 cells were seeded in the upper chamber and the same number of macrophages with/without lincRNA-p21 knockdown were put in lower chamber and co-cultured at 37 °C. After 24 h, 4T1 cells were fixed with 4% paraformaldehyde, then stained with crystal violet. The number of migrated cells was counted using a phase-contrast microscope.
Invasion was measured by wound healing assay. 5 × 104 4T1 cells were seeded in the upper chamber and the same number of macrophages with/without lincRNA-p21 knockdown were put in lower chamber and co-cultured at 37 °C. After 48 h, the upper chamber was removed and a small pipette tip was used to make a linear wound across the confluent cell layer and cultured for 24 h. Cells were rinsed twice to remove wreckage and detached cells and the dimension of wound were observed.
RNA fluorescence in-situ hybridization
Cell growth in 24-well plates reached 60%–70% confluence on coverslips, washed for 5 min with PBS, then fixed in 4% paraformaldehyde for 10 min and washed for 5 min with PBS for three times. Cells were permeabilized in PBS containing 0.5% Triton X-100 at 4 °C for 5 min and again washed for 5 min with PBS three times. After blockade, hybridization was performed overnight at 37 °C in hybridization buffer containing lncRNA FISH Probe Mix stock solution (Ribobio, Guangzhou, China). 4 × SSC (containing 0.5% Tween-20) was used to wash cells three times; subsequently, 2 × SSC and 1 × SSC were used, respectively. Celluar nulcear was stained by DAPI, the images were carptured by Zeiss LSM 510 Meta Laser Scanning Confocal Microscope.
Statistical analysis
GraphPad Prism 5 was used for all statistical analysis. The data were presented as the mean ± SEM from at least three independent experiments. Student’s unpaired t-test and one-way ANOVA were performed to compare the differences between two or multiple groups. A log-rank (Mantel-Cox) was used to analyze the survival time of tumor-bearing mice. P < 0.05 was considered as statistically significant.
Results
LincRNA-p21 was significantly up-regulated in TAMs
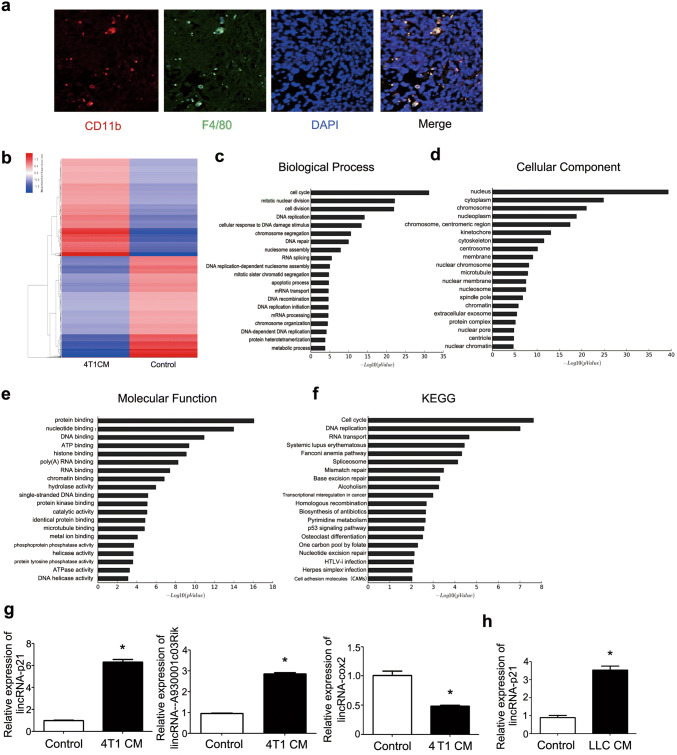
More and more data showed that macrophage infiltration was associated with cancer development and metastasis [27]. To demonstrate the crosstalk between TAMs and breast cancer, the macrophage infiltration was detected in MMTV-PyVT mice. As shown in Fig. 1a, most TAMs existed in primary tumor tissue. To explore the function of TAMs under breast cancer milieu, the lncRNAs microarray analysis was used to detect the changes of lncRNAs between normal and treated macrophage by 4T1CM. The results showed that 305 and 697 lncRNAs were significantly up- and down-regulated, respectively (Fig. 1b). GO analysis revealed that the up- and down-regulated genes were enriched in cell cycle, apoptotic process and p53 signaling pathway (Fig. 1c–f). Some of lncRNA changes were confirmed and consistent with the results of microarray. LincRNA-p21 was significantly up-regulated (Fig. 1g). Additionally, lincRNA-p21 change was also detected in other tumor conditioned medium educated macrophages such as LLC and the similar phenomena were detected (Fig. 1h).
Fig. 1.
LincRNA-p21 up-regulated in TAMs. a TAMs infiltrated in tumor tissue. Representative immunofluorescence microscopy images were shown. Red, CD11b; Green, F4/80; Blue, nuclei. 5 MMTV-PyVT mice were included. b Heat map of lncRNAs profiles in TAMs. Macrophages were treated by 4T1 cultured supernatant (4T1CM) for 48 h compared with control macrophages. Red and blue mean up-regulation and down-regulation, respectively. c–e GO analysis was performed to describe their functions: c biological processes, d cellular components and e molecular functions. f KEGG database was used to analyze pathway, and the significance of differentially expressed genes’ enrichment in each pathway entry. g The expressions of lincRNA-p21, lincRNA-cox2 and lncRNA-A930001c03Rik were detected by RT-qPCR. Macrophages were treated by 4T1CM for 48 h and then collected using for RT-qPCR analysis. H LincRNA-p21 expression increased in LLC supernatant-treated macrophages. Data were obtained from three independent experiments and the representative images shown. *P < 0.05. LLC mean Lewis lung carcinoma cells
LincRNA-p21 knockdown reversed the functional phenotype of TAMs
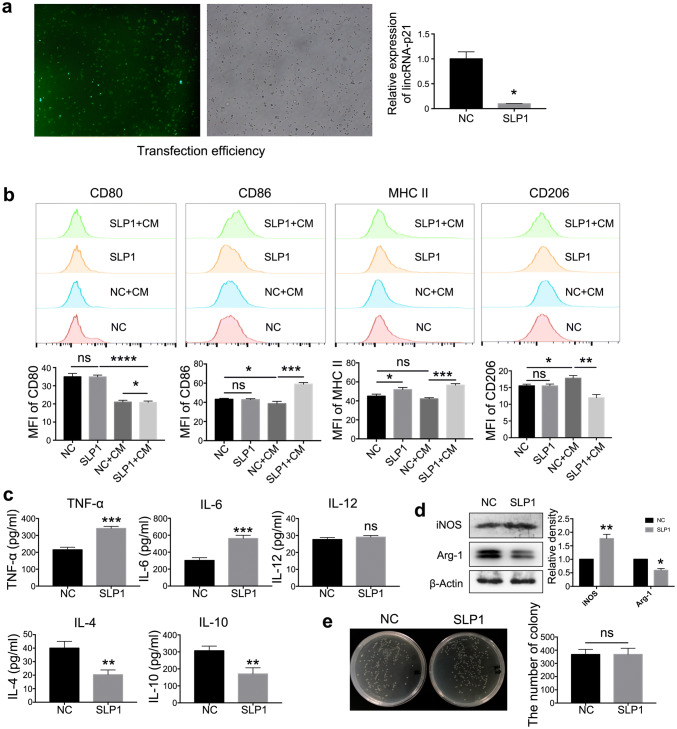
To confirm the effects of lincRNA-p21 on macrophage function in tumor milieu, siRNA was employed to down-regulate the expression of lincRNA-p21. The transfection efficiency and knockdown ability of siRNA were shown on Fig. 2a. LincRNA-p21 knockdown up-regulated CD86 and MHCII and down-regulated CD206; however, there was no effect on CD80 (Fig. 2b). Additionally, lincRNA-p21 knockdown benefited the pro-inflammatory mediator production such as IL-6 and TNF-α and inhibited the anti-inflammatory factors such as IL-4 and IL-10 secretion;; conversely, IL-12 production was no different between the two groups (Fig. 2c).
Fig. 2.
LincRNA-p21 knockdown reversed the functional phenotype of TAMs in tumor milieu. a The transfection efficiency reached 96% (left) and the relative expression of lincRNA-p21 (right). Fluorescent labeled siRNAs were designed to down-regulate lincRNA-p21 in macrophages. The transfection efficiency was observed under fluorescence microscope after 6 h, lincRNA-p21 expression was detected by RT-qPCR after 24 h. Data are presented as mean ± SEM. b CD86 and MHC II up-regulated; conversely, CD206 down-regulated following lincRNA-p21 knockdown in TAMs. The upper panels showed representative images and lower panels showed statistical analysis. c LincRNA-p21 knockdown promoted pro-inflammatory cytokines’ secretion of TAMs. Secretion of IL-4, IL-6, IL-10, IL-12 and TNF-α was assessed by ELISA in the supernatant. d The levels of iNOS up-regulated; however, Arg-1 down-regulated following lincRNA-p21 knockdown in TAMs. Western blotting analysis was employed to assess the levels of iNOS and Arg-1. β-actin was used as a loading control. The left panels show representative blots and right panels show statistical analysis. e LincRNA-p21 knockdown could not increase phagocytosis of TAMs. LincRNA-p21 was knockdown in TAMs, then Escherichia coli (E. coli) was added. After 1 h, the bacteria were sterilized, TAMs were lysed. The number of bacteria phagocytized by TAMs was counted. The left panels show representative images and right panels show statistical analysis. Data are presented as mean ± SEM. All the data were obtained from three independent experiments and the representative images are shown. *P < 0.05 relative to control. N.S. means no significance. SLP1 means lincRNA-p21 downregulated TAMs
It is well known that M2 macrophages promote the proliferation of tumor cells by I-arginine as the substrate to synthesize ornithine and polyamines [28]. Under normal circumstances, the expression and activity of iNOS and Arg-1 were strictly regulated in macrophages, and the dynamic balance plays an important role in maintaining the functional stability of macrophages. LincRNA-p21 knockdown resulted in the increase of iNOS and decrease of Arg-1 in macrophages following 4T1CM treatment (Fig. 2d). Conversely, the phagocytic function did not have any difference (Fig. 2e). These observations suggested that lincRNA-p21 knockdown in macrophages could increase the proportion of M1 macrophages and decrease the proportion of M2 macrophages, which benefited the anti-tumor function.
LincRNA-p21 down-regulated TAMs promoted tumor cell apoptosis and inhibited their migration and invasion
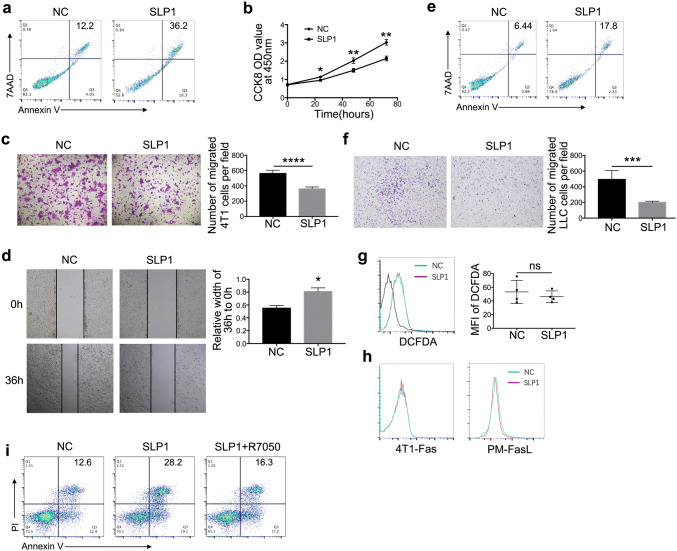
To investigate the effect of lincRNA-p21 down-regulated TAMs on tumor cells, lincRNA-p21 down-regulated TAMs were co-cultured with 4T1 cells. LincRNA-p21 down-regulated TAMs promoted cancer cells apoptosis and inhibited their proliferation, migration and invasion (Fig. 3a–d). The similar phenomena were also observed in LLC (Fig. 3e, f). However, it remains unclear about lincRNA-p21 down-regulated TAMs inducing tumor cells apoptosis. Previous studies have shown that macrophages kill tumor cells dependent on ROS production [29]. Therefore, ROS expression was detected. As Fig. 3g shows, there are no significant differences between lincRNA-p21 down-regulated and control macrophages. We try to figure out whether the apoptosis induced by lincRNA-p21 down-regulated TAMs was associated with Fas-FasL pathway. FasL and Fas expression on lincRNA-p21 down-regulated TAMs and tumor cells were detected, respectively. There was also no obvious difference (Fig. 3h). But TNF-α level was up-regulated following lincRNA-p21 knockdown in TAMs (Fig. 2c). We hypothesized whether apoptosis of tumor cells was caused by increase of TNF-α secretion in lincRNA-p21 down-regulated TAMs; therefore, 4T1 cells were preteated by R7050, TNF-α receptor antagonist, and then co-cultured with lincRNA-p21 down-regulated TAMs, the results showed that the apoptosis induced by lincRNA-p21 down-regulated TAMs was blocked. The result further confirmed our speculation (Fig. 3i).
Fig. 3.
LincRNA-p21 down-regulated TAMs promoted tumor cells apoptosis, inhibited their migration and invasion. a, e LincRNA-p21 down-regulated TAMs promoted tumor cell apoptosis. 4T1 and LLC were co-cultured with lincRNA-p21 down-regulated macrophages for 48 h, respectively. The tumor cells were collected and stained with Annexin-V and 7AAD to detect the apoptosis using flow cytometry, respectively. b LincRNA-p21 down-regulated TAMs inhibited 4T1 cell proliferation. The proliferation of 4T1 cells co-cultured with lincRNA-p21 down-regulated macrophages was measured using Cell Counting Kit-8 (CCK-8) at indicated point. c, f LincRNA-p21 down-regulated TAMs inhibited tumor cells migration. Transwell assays were performed to detect migration ability of tumor cells (4T1 and LLC) co-cultured with lincRNA-p21 down-regulated TAMs for 24 h. The left panels show representative images and right panels show statistical analysis. d LincRNA-p21 down-regulated TAMs inhibited 4T1 cells invasion. Wound-healing assay was used to detect the invasive ability of 4T1 cells co-cultured with lincRNA-p21 down-regulated TAMs. g LincRNA-p21 knockdown could not increase ROS production in TAMs. The left panels show representative images and right panels show statistical analysis. h The expression of Fas on 4T1 cell and FasL on TAMs was assessed by flow cytometry. i R7050 (10 nM) was used to pretreat 4T1 for 6 h and after that 4T1 was co-cultured with lincRNA-p21 down-regulated macrophages for 48 h; 4T1 cells were collected and stained with Annexin V and PI to detect apoptosis. All the data were obtained from three independent experiments and the representative images are shown. *P<0.05 relative to control. N.S. means no significance. LLC and SLP1 mean Lewis lung carcinoma cells and lincRNA-p21 downregulated macrophages
LincRNA-p21 knockdown inducing functional reversion of TAMs may be caused by promoting MDM2 to antagonize p53 function
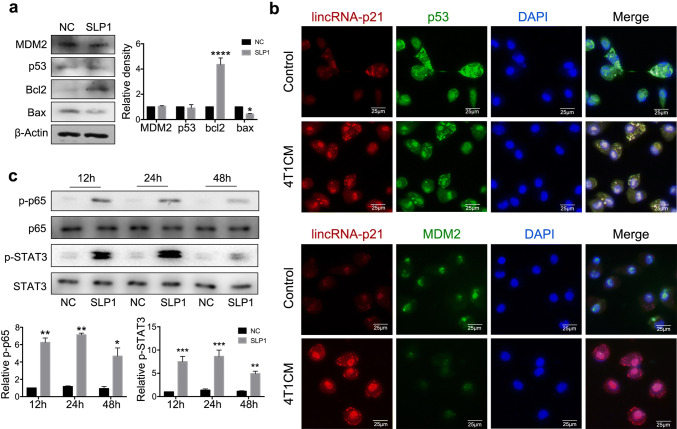
Published data have demonstrated that lincRNA-p21 regulates p53-dependent target genes, Bax, Puma and Noxa without altering p53 expression level [23, 30]. MDM2, an E3 ubiquitin ligase, antagonizes p53 by enhancing its proteasome-dependent degradation. Therefore, Bax, Bcl-2, p53 and MDM2 protein levels were detected in lincRNA-p21 down-regulated TAMs, Bax was down-regulated and Bcl-2 was up-regulated compared with control; however, there were no obvious difference about p53 and MDM2 expression (Fig. 4a). To further confirm the potential regulations among lincRNA-p21, p53 and MDM2, lincRNA-p21 was labeled with FISH probe, MDM2 and p53 proteins were detected by immunofluorescence, respectively, in TAM, and the results showed that lincRNA-p21 and p53 are co-localized in cytoplasm; conversely, MDM2 mainly localized in the nucleus membrane and its expression was decreased (Fig. 4b). Therefore, we speculated that in tumor milieu, the increasing lincRNA-p21 abolished MDM2 degradation to p53. LincRNA-p21 knockdown, MDM2 binds to p53 and elicits p53 degradation, which promotes TAMs to show a pro-inflammatory M1 phenotype. Previous data showed that activation of NF-κB and STAT3 promotes the macrophages’ polarization into M1 phenotype [31]. To confirm lincRNA-p21 knockdown inducing functional reversion of TAMs dependent on the above process, NF-κB-STAT3 pathway was detected in lincRNA-p21 down-regulated TAMs. As shown in Fig. 4c, NF-κB and STAT3 were activated at 24 h. Taken together, lincRNA-p21 knockdown in TAMs might facilitate MDM2 to antagonize p53 activation, activate NF-κB and STAT3 signaling pathways and reverse TAM phenotype in tumor milieu.
Fig. 4.
LincRNA-p21 knockdown inducing functional reversion of TAMs may be caused by promoting MDM2 to antagonize p53 function. a LincRNA-p21 knockdown increased Bcl-2 and decreased Bax expression in TAMs. The left panels show representative blots and right panels show statistical analysis. b LincRNA-p21 and p53 are co-localized in cytoplasm; conversely, MDM2 localized in the nucleus membrane. LincRNA-p21 was labeled with FISH probe and hybridized in TAMs; MDM2 and p53 expressions were detected by immunofluorescence. The nuclei were stained with DAPI. c LincRNA-p21 knockdown activated STAT3 and p65 in TAMs. Macrophages with/without lincRNA-p21 knockdown were treated by 4T1CM and cells were collected at indicated points for using western blotting analysis. The upper panels show representative blots and lower panels show statistical analysis. All data were obtained from three independent experiments and the representative images are shown. SLP1 means lincRNA-p21 downregulated macrophages
LincRNA-p21 knockdown macrophages adoptive transfer could alleviate breast cancer development
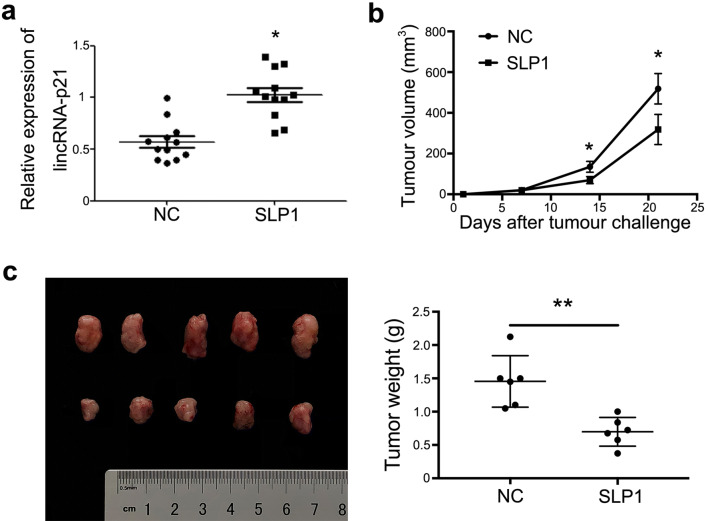
To demonstrate tumorigenesis of lincRNA-p21 down-regulated macrophages in vivo, TAMs were isolated from breast cancer-bearing mice to detect the expression of lincRNA-p21, and the same mice spleen macrophages were used as control. The results showed that the expression of lincRNA-p21 in macrophages from tumor tissues was significantly up-regulated compared with control (Fig. 5a).
Fig. 5.
LincRNA-p21 knockdown macrophages’ adoptive transfer could alleviate breast cancer development. 1 × 106 4T1 cells and 1 × 105 primary macrophages with/without lincRNA-p21 knockdown were subcutaneously (s.c.) inoculated into the backside of 10 BALB/c mice to generate tumors. After tumor inoculation, 1 × 105 macrophages/week with/without lincRNA-p21 knockdown were injected into the solid tumor until the mice were killed. a LincRNA-p21 expression was increased in macrophages from tumors by RT-qPCR. Every dot represents one mouse. b LincRNA-p21 down-regulated macrophages decreased the tumor volumes. c LincRNA-p21 down-regulated macrophages decreased the tumor weights. Data are presented as mean ± SEM. *P<0.05 relative to control
Next, 4T1 and primary macrophages, 4T1 and primary macrophages with lincRNA-p21 knockdown were inoculated into mice to construct subcutaneous tumors, respectively. After inoculation, the primary macrophages with/without lincRNA-p21 knockdown were injected into tissue one time/week until the mice were killed, the volumes of tumors were measured every 3 days. The results showed that lincRNA-p21 knockdown macrophages alleviated the tumor development, decreased tumor volumes and weights compared with control group (Fig. 5b, c). Furthermore, the survival time was dramatically extended in lincRNA-p21 knockdown macrophages injection group (Fig. 5d). All these data suggested that lincRNA-p21 expression up-regulated in TAMs and lincRNA-p21 knockdown macrophages adoptive transfer could alleviate breast cancer progression.
Discussion
Macrophages are highly heterogeneous phagocytic population and play critical roles in inflammatory development, wound healing, tissue repair and regeneration as well as cancer progression [32]. Exposure to different stimulus induces macrophage differentiation/polarization into: classically activated M1 and alternatively activated M2. M1 macrophages inhibit tumor progression through expression of pro-inflammatory factors, such as TNF-α. However, during tumor development, macrophages differentiate/polarize into M2 phenotype, namely TAMs, and secrete IL-4 and IL-10, enhance angiogenesis, matrix synthesis and immune escape [33–35]. In breast cancer microenvironment, TAMs are critical tumor-promoting cells, for example, promoting breast cancer cell proliferation, invasion, metastasis and immune escape, or contributing to resistance to multiple drugs. TAMs are also correlated with a worse prognosis of breast cancer [36]. Therefore, it was critical to classify the maintenance mechanisms of specific TAMs phenotype in breast cancer microenvironment.
LincRNA p21 has been reported in some cancers, but it is believed to play a role in cancer cells; however, how lincRNA p21 works in immune cells and how lincRNA p21 is regulated in TAMs in breast cancer have not been defined. We speculate that this may be related to some cytokines released during tumor development, and the specific mechanism needs to be further explored. In the present study, we demonstrated that lincRNA-p21 knockdown significantly reversed the functional phenotype of TAMs and increased their anti-tumor ability. LincRNA-p21 could directly target on p53, or indirectly target on p53 by hnRNP-K dependent mechanism. LincRNA-p21 binds to MDM2 and promotes P300 interaction with p53, thereby benefiting p53 acetylation and increasing its activity [37–39]. However, our results showed that lincRNA-p21 directly targeted on p53, abolished MDM2 degradation to p53 and facilitated phenotype maintenance of TAMs in breast cancer microenvironment; conversely, lincRNA-p21 knockdown promoted MDM2 degradation to p53, formed NF-κB-IL-6-STAT3 loop [40] and reversed functional phenotype of TAMs [41], which was consistent with previous data that p53/MDM2 complex regulates the polarization of macrophage [42]. Therefore, we speculated that lincRNA-p21 biological function is induced by different stimuli and lincRNAp-21 plays multiple roles by binding to different proteins. LincRNA-p21 may be a critical determinant factor of p53/MDM2 stability. Of course, the mechanisms are needed to further confirm whether lincRNA-p21 also selectively binds to hnRNP-K.
The lincRNA-p21 down-regulated macrophages did not affect their phagocytosis but promoted tumor cells apoptosis, migration and invasion. However, it is unclear whether lincRNA-p21 down-regulated macrophages can kill tumor cells by recruiting CTLs and NK cells to the tumor site. Furthermore, lincRNA-p21 down-regulated macrophages could delay the occurrence of tumors at the early stages of tumor inoculationand prolong the survival time of tumor-bearing mice. LincRNA-p21, an evolutionarily conserved gene, is also expressed in the human genome [43]. It is unclear whether lincRNA-p21 also changes in tumor patient’s macrophages and whether lincRNA-p21 can be used as a prognostic indicator or an early predictor for the diagnosis of tumors. However, our data at least indicated that lincRNA-p21 was an important regulator to the domestication of infiltrated monocytes/macrophages in tumor microenvironment.
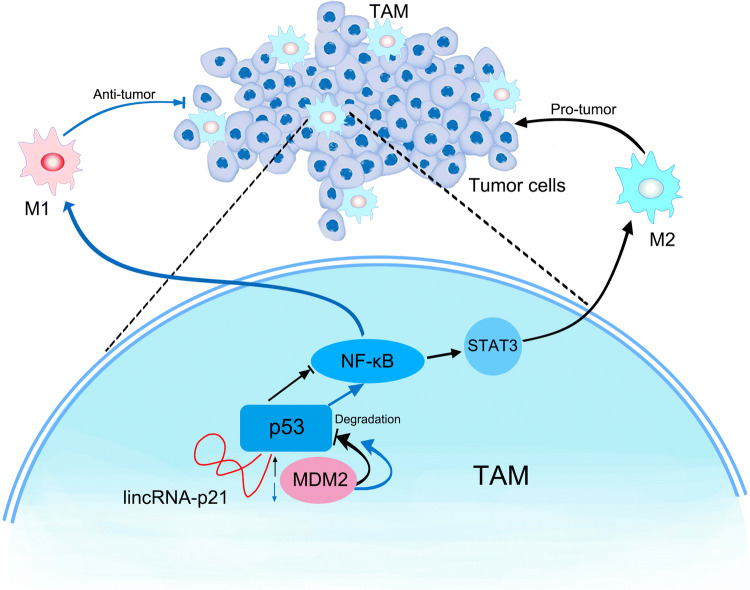
In conclusion, under tumor microenvironment, monocytes/macrophages infiltrate into the tumorigenic site and are domesticated into TAMs by enhancing lincRNA-p21 expression and promoting its interaction with p53 and contribute to the tumor development. LincRNA-p21 knockdown resulted in the reduction between p53 and lincRNA-p21 interaction, facilitated p53 interaction with MDM2, induced NF-κB and STAT3 signaling pathway activation, reversed TAMs phenotype, produced TNF-α to kill tumor cells and thereby exerted anti-tumor function (Fig. 6). Our data indicated that lincRNA-p21 was an important regulator of TAM function in tumor microenvironment. Our data also suggested that lincRNA-p21 may be a novel therapeutic target of cancers characterized by macrophages’ infiltration.
Fig. 6.
Schematic of LincRNA-p21 knockdown reverses TAMs function and alleviates breast cancer development
Acknowledgements
This work was supported by Key University Science Research Project of Jiangsu Province (Grant No. 16KJA320005), the Social Development Foundation of Jiangsu Province (Grant No. BE2016716). The maternal and child project in Jiangsu Province (Grant No. F201511); Changzhou applied basic research project (CJ20180037).
Abbreviation
- ACK
Red blood cell lysis buffer
- CCK-8
Cell counting kit-8
- CTL
Cytotoxic T lymphocyte
- GO
Gene ontology
- LLC
Lewis lung carcinoma
- lncRNA
Long non-coding RNA
- miRNA
Small RNA
- NK
Natural killer
- ROS
Reactive oxygen species
- siRNA
Small interfering RNA
- SPF
Specific pathogen-free
- TAMs
Tumor-associated macrophages
- 4T1CM
4T1 mouse breast tumor cell supernatant
Author contributions
LZ, YT and FG did most experiment, prepared Figs. 1, 2, 3 and 4, BY and JL prepared Figs. 5 and 6, ZS and HX designed the project, ZS prepared the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that there is no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Lining Zhou, Yu Tian and Fang Guo contributed equally to this work.
References
- 1.DeSantis C, Ma J, Bryan L. Jemal A (2014) Breast cancer statistics. CA-Cancer J Clin. 2013;64(1):52–62. doi: 10.3322/caac.21203. [DOI] [PubMed] [Google Scholar]
- 2.Huneidi SA, Wright NC, Atkinson A, Bhatia S, Singh P. Factors associated with physical inactivity in adult breast cancer survivors-A population-based study. Cancer Med. 2018 doi: 10.1002/cam4.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matsumoto A, Jinno H, Ando T, Fujii T, Nakamura T, Saito J, Takahashi M, Hayashida T, Kitagawa Y. Biological markers of invasive breast cancer. Jpn J Clin Oncol. 2016;46(2):99–105. doi: 10.1093/jjco/hyv153. [DOI] [PubMed] [Google Scholar]
- 4.King TA, Morrow M. Surgical issues in patients with breast cancer receiving neoadjuvant chemotherapy. Nat Rev Clin Oncol. 2015;12(6):335–343. doi: 10.1038/nrclinonc.2015.63. [DOI] [PubMed] [Google Scholar]
- 5.Soysal SD, Tzankov A, Muenst SE. Role of the tumor microenvironment in breast cancer. Pathobiology. 2015;82(3–4):142–152. doi: 10.1159/000430499. [DOI] [PubMed] [Google Scholar]
- 6.Bussard KM, Mutkus L, Stumpf K, Gomez-Manzano C, Marini FC. Tumor-associated stromal cells as key contributors to the tumor microenvironment. Br Cancer Res. 2016;18(1):84. doi: 10.1186/s13058-016-0740-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goswami KK, Ghosh T, Ghosh S, Sarkar M, Bose A, Baral R. Tumor promoting role of anti-tumor macrophages in tumor microenvironment. Cell Immunol. 2017;316:1–10. doi: 10.1016/j.cellimm.2017.04.005. [DOI] [PubMed] [Google Scholar]
- 8.Murray PJ. Macrophage polarization. Annu Rev Physiol. 2017;79:541–566. doi: 10.1146/annurev-physiol-022516-034339. [DOI] [PubMed] [Google Scholar]
- 9.Liu YC, Zou XB, Chai YF, Yao YM. Macrophage polarization in inflammatory diseases. Int J Biol Sci. 2014;10(5):520–529. doi: 10.7150/ijbs.8879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tariq M, Zhang J, Liang G, Ding L, He Q, Yang B. Macrophage polarization: anti-cancer strategies to target tumor-associated macrophage in breast cancer. J Cell Biochem. 2017;118(9):2484–2501. doi: 10.1002/jcb.25895. [DOI] [PubMed] [Google Scholar]
- 11.Mills CD, Lenz LL, Harris RA. A breakthrough: macrophage-directed cancer immunotherapy. Cancer Res. 2016;76(3):513–516. doi: 10.1158/0008-5472.can-15-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ngambenjawong C, Gustafson HH, Pun SH. Progress in tumor-associated macrophage (TAM)-targeted therapeutics. Adv Drug Deliv Rev. 2017;114:206–221. doi: 10.1016/j.addr.2017.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dehne N, Mora J, Namgaladze D, Weigert A, Brune B. Cancer cell and macrophage cross-talk in the tumor microenvironment. Curr Opin Pharmacol. 2017;35:12–19. doi: 10.1016/j.coph.2017.04.007. [DOI] [PubMed] [Google Scholar]
- 14.Aras S, Zaidi MR. TAMeless traitors: macrophages in cancer progression and metastasis. Br J Cancer. 2017;117(11):1583–1591. doi: 10.1038/bjc.2017.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mantovani A, Marchesi F, Malesci A, Laghi L, Allavena P. Tumour-associated macrophages as treatment targets in oncology. Nat Rev Clin Oncol. 2017;14(7):399–416. doi: 10.1038/nrclinonc.2016.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woo SR, Corrales L, Gajewski TF. Innate immune recognition of cancer. Annu Rev Immunol. 2015;33:445–474. doi: 10.1146/annurev-immunol-032414-112043. [DOI] [PubMed] [Google Scholar]
- 17.Biswas SK, Mantovani A. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat Immunol. 2010;11(10):889–896. doi: 10.1038/ni.1937. [DOI] [PubMed] [Google Scholar]
- 18.Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29(4):452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferre F, Colantoni A, Helmer-Citterich M. Revealing protein-lncRNA interaction. Brief Bioinform. 2016;17(1):106–116. doi: 10.1093/bib/bbv031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marchese FP, Raimondi I, Huarte M. The multidimensional mechanisms of long noncoding RNA function. Genome Biol. 2017;18(1):206. doi: 10.1186/s13059-017-1348-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen YG, Satpathy AT, Chang HY. Gene regulation in the immune system by long noncoding RNAs. Nat Immunol. 2017;18(9):962–972. doi: 10.1038/ni.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fischer M. Census and evaluation of p53 target genes. Oncogene. 2017;36(28):3943–3956. doi: 10.1038/onc.2016.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu G, Cai J, Han Y, Chen J, Huang ZP, Chen C, Cai Y, Huang H, Yang Y, Liu Y, Xu Z, He D, Zhang X, Hu X, Pinello L, Zhong D, He F, Yuan GC, Wang DZ, Zeng C. LincRNA-p21 regulates neointima formation, vascular smooth muscle cell proliferation, apoptosis, and atherosclerosis by enhancing p53 activity. Circulation. 2014;130(17):1452–1465. doi: 10.1161/circulationaha.114.011675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Castellano JJ, Navarro A, Vinolas N, Marrades RM, Moises J, Cordeiro A, Saco A, Munoz C, Fuster D, Molins L, Ramirez J, Monzo M. LincRNA-p21 impacts prognosis in resected non-small cell lung cancer patients through angiogenesis regulation. J Thorac Oncol. 2016;11(12):2173–2182. doi: 10.1016/j.jtho.2016.07.015. [DOI] [PubMed] [Google Scholar]
- 25.Spurlock CF, 3rd, Tossberg JT, Matlock BK, Olsen NJ, Aune TM. Methotrexate inhibits NF-kappaB activity via long intergenic (noncoding) RNA-p21 induction. Arthritis Rheumatol. 2014;66(11):2947–2957. doi: 10.1002/art.38805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen Y, Liu Y, Sun T, Yang W. LincRNA-p21 knockdown enhances radiosensitivity of hypoxic tumor cells by reducing autophagy through HIF-1/Akt/mTOR/P70S6K pathway. Exp Cell Res. 2017;358(2):188–198. doi: 10.1016/j.yexcr.2017.06.016. [DOI] [PubMed] [Google Scholar]
- 27.Na YR, Je S, Seok SH. Metabolic features of macrophages in inflammatory diseases and cancer. Cancer Lett. 2018;413:46–58. doi: 10.1016/j.canlet.2017.10.044. [DOI] [PubMed] [Google Scholar]
- 28.Artyomov MN, Sergushichev A, Schilling JD. Integrating immunometabolism and macrophage diversity. Semin Immunol. 2016;28(5):417–424. doi: 10.1016/j.smim.2016.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tan HY, Wang N, Li S, Hong M, Wang X, Feng Y. The reactive oxygen species in macrophage polarization: reflecting its dual role in progression and treatment of human diseases. Oxid Med Cell Longev. 2016;2016:2795090. doi: 10.1155/2016/2795090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wade M, Li YC, Wahl GM. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat Rev Cancer. 2013;13(2):83–96. doi: 10.1038/nrc3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jin X, Yao T, Zhou Z, Zhu J, Zhang S, Hu W, Shen C. Advanced glycation end products enhance macrophages polarization into M1 phenotype through activating RAGE/NF-kappaB pathway. Biomed Res Int. 2015;2015:732450. doi: 10.1155/2015/732450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luo Z, Wang Q, Lau WB, Lau B, Xu L, Zhao L, Yang H, Feng M, Xuan Y, Yang Y, Lei L, Wang C, Yi T, Zhao X, Wei Y, Zhou S. Tumor microenvironment: the culprit for ovarian cancer metastasis? Cancer Lett. 2016;377(2):174–182. doi: 10.1016/j.canlet.2016.04.038. [DOI] [PubMed] [Google Scholar]
- 33.Choi J, Gyamfi J, Jang H, Koo JS. The role of tumor-associated macrophage in breast cancer biology. Histol Histopathol. 2018;33(2):133–145. doi: 10.14670/HH-11-916. [DOI] [PubMed] [Google Scholar]
- 34.Sica A, Erreni M, Allavena P, Porta C. Macrophage polarization in pathology. Cell Mol Life Sci. 2015;72(21):4111–4126. doi: 10.1007/s00018-015-1995-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rothlin CV, Carrera-Silva EA, Bosurgi L, Ghosh S. TAM receptor signaling in immune homeostasis. Annu Rev Immunol. 2015;33:355–391. doi: 10.1146/annurev-immunol-032414-112103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qiu SQ, Waaijer SJH, Zwager MC, de Vries EGE, van der Vegt B, Schroder CP. Tumor-associated macrophages in breast cancer: innocent bystander or important player? Cancer Treat Rev. 2018;70:178–189. doi: 10.1016/j.ctrv.2018.08.010. [DOI] [PubMed] [Google Scholar]
- 37.Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, Attardi LD, Regev A, Lander ES, Jacks T, Rinn JL. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142(3):409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Inoue K, Fry EA, Frazier DP. Transcription factors that interact with p53 and Mdm2. Int J Cancer. 2016;138(7):1577–1585. doi: 10.1002/ijc.29663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang Z, Chen WY, Shimada M, Nguyen UT, Kim J, Sun XJ, Sengoku T, McGinty RK, Fernandez JP, Muir TW, Roeder RG. SET1 and p300 act synergistically, through coupled histone modifications, in transcriptional activation by p53. Cell. 2013;154(2):297–310. doi: 10.1016/j.cell.2013.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fan Y, Mao R, Yang J. NF-kappaB and STAT3 signaling pathways collaboratively link inflammation to cancer. Protein Cell. 2013;4(3):176–185. doi: 10.1007/s13238-013-2084-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo H, Jin D, Chen X. Lipocalin 2 is a regulator of macrophage polarization and NF-kappaB/STAT3 pathway activation. Mol Endocrinol. 2014;28(10):1616–1628. doi: 10.1210/me.2014-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li L, Ng DS, Mah WC, Almeida FF, Rahmat SA, Rao VK, Leow SC, Laudisi F, Peh MT, Goh AM, Lim JS, Wright GD, Mortellaro A, Taneja R, Ginhoux F, Lee CG, Moore PK, Lane DP. A unique role for p53 in the regulation of M2 macrophage polarization. Cell Death Differ. 2015;22(7):1081–1093. doi: 10.1038/cdd.2014.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tang SS, Zheng BY, Xiong XD. LincRNA-p21: Implications in Human Diseases. Int J Mol Sci. 2015;16(8):18732–18740. doi: 10.3390/ijms160818732. [DOI] [PMC free article] [PubMed] [Google Scholar]