Dear Editors,
We recently published the tool MuPeXI, the mutant peptide extractor and informer, enabling neoepitope prediction from tumor sequencing data [1]. MuPeXI is originally designed for variant calls obtained from sequencing data of human origin but increasing interest to determine neoepitopes in murine models have encouraged us to update and test MuPeXI for mouse compatibility. The murine-compatible MuPeXI is now available as a command line tool (https://github.com/ambj/MuPeXI) together with a mouse-specific web server (http://www.cbs.dtu.dk/services/MuPeXI-mouse/).
Despite the interest for determining neoepitopes from preclinical mouse models, only few tools for neoepitope prediction have been designed and evaluated to allow neoepitope prediction from data of murine origin. To fulfill this need, we optimized MuPeXI to enable identification of murine neopeptides. MuPeXI is now compatible with the genetic reference of mus musculus, as well as the two commonly used mouse strains, BALBc and C57BL/6. To test the NGS pipeline and optimize MuPeXI, we evaluate the neoepitope landscape in the CT26 tumor cell line, which has been extensively used in mode-of-action studies in syngeneic mouse tumor models [2], and proven especially valuable as an experimental model for immune therapy interventions. We used sequencing data from Castle et al. [2] and Mosely et al. [3], including both the CT26 cell line (CL) and the CT26 tumors grown in vivo on BALBc mice (TU). The NGS analysis pipeline was followed as suggested by Genome Analysis Tool Kit (GATK), best practice guidelines, using the same tools as in the original MuPeXI paper [1]. References were downloaded from Ensembl’s mouse genome assemble, further detail can be found in the MuPeXI user manual (https://github.com/ambj/MuPeXI/blob/master/doc/MuPeXI_User_Manual.md#references). In the analysis we incorporated the new binding predictor netH2pan into MuPeXI. NetH2pan is trained solely on mouse-binding affinity and eluted ligand data [4], thereby providing the most suitable H2-binding predictions for the neopeptides extracted from somatic variant of murine sequencing data by MuPeXI.
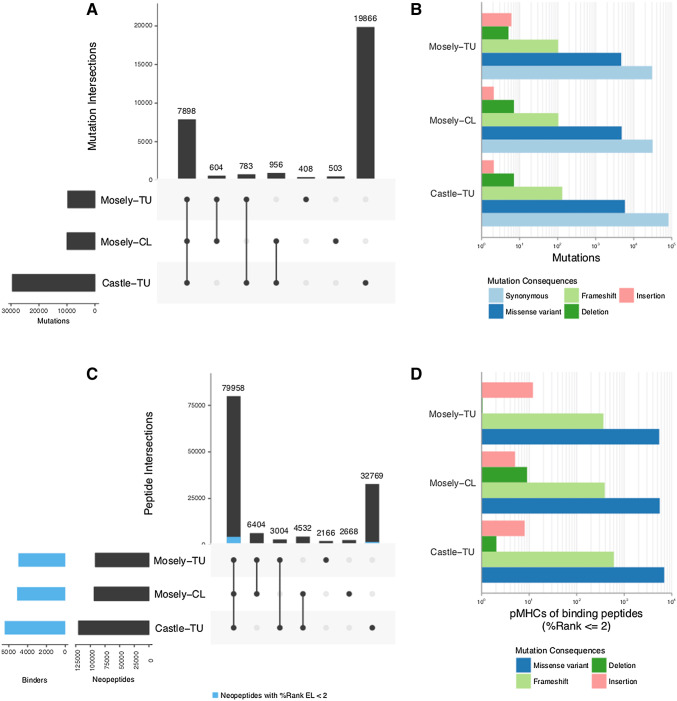
The mutational landscape was compared for the three samples tested, and although the total number of mutations identified in the three samples were high, it was in accordance with the original papers. The analysis revealed that only a fraction of the mutations, 7898 (~ 26% of the total) was identified in all 3 samples (Fig. 1a). The number of non-synonymous mutations (NSmut) did not vary substantially compared to the total number of mutations identified, i.e., Castle: 5994–3.8%, Mosely CL: 4921–3.1%, Mosely TU: 4793–3.0% (Fig. 1b). Besides missense variant (MV) mutations, frame shifts, and indels (FI) were also identified, but present to a lesser degree (MV: 15349–9.6%, FI: 359–0.2%). To identify how many of the NSmuts lead to potential neoepitopes, the updated murine-compatible MuPeXI software was used with the relevant BALBc references. We identified 79,958 (61% of all) neopeptides shared among the two tumor samples obtained from two different studies and the original CT26 cell line sample (Fig. 1c). Of these, 4399 (3.4%) had an eluted ligand percentile rank score (%Rank EL) below 2 and were considered binding peptides in all 3 samples (Fig. 1c, blue). A total of 7034 (5.4%) peptides were identified as binders and therefore potential neoepitopes out of the total 124,467 unique neopeptides extracted from all 3 samples. Of the binding peptides, 61 potential minimal neopeptides matched previously described long peptides shown to evoke immune responses [5]. The potential neoepitopes originate from various types of NSmuts, including frameshifts and indels (Fig. 1d). Importantly, frameshift mutations translate to numerous predicted neopeptides per mutation and consequently, frameshift mutations contribute to 2.1% for the total NSmuts, while 7% of the predicted neopeptides, therefore it will be interesting to investigate the true immunogenicity of frameshift neopeptides in CT26. With the murine-compatible version of MuPeXI, we were able to show that 87% of predicted potential neoepitopes is identical between the CT26 tumor cell line and the established tumor originating from the same batch. Furthermore, across different laboratories, established tumor of the CT26 cell line showed 67% overlap in predicted potential neoepitopes. The murine-compatible version of MuPeXI together with the tested NGS pipeline gives the user an easy way to extract neopeptides from murine sequencing data and evaluate their immunogenicity potential.
Fig. 1.
CT26 neopeptide and mutation landscape. a Overlap in predicted neopeptides of 3 individual samples (from 2 studies, Castle et al. and Mosely et al. including 2 established CT26 tumors and 1 CT26 cell line). Peptides predicted as binders by netH2pan with an eluted ligand percentile rank score (%Rank EL) below 2 is indicated in blue. b Overlap in all mutations identified by MuTect2-GATK3.7 and passing the default filters. c Number of mutations for individual mutation consequences determined by Ensembl’s variant effect predictor (VEP). d Number of peptide MHC complexes (pMHCs) of peptides originating from individual mutation consequences determined by VEP. CL cell line, TU tumor
As we highlighted in the original MuPeXI paper, additional data is needed to optimize the prediction tools for determination of the immunogenicity of neopeptides [1]. Because murine models provide a more readily available experimental setting than human patients, the application of MuPeXI in a murine setting could help produce such data, to improve the prediction model on large amounts of murine data prior to testing and validation on human data.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Anne-Mette Bjerregaard and Thomas Kainamura Pedersen contributed equally.
Contributor Information
Anne-Mette Bjerregaard, Phone: +45 35886290, Email: ambj@dtu.dk.
Sine Reker Hadrup, Email: sirha@dtu.dk.
References
- 1.Bjerregaard A-M, Nielsen M, Hadrup SR, et al. MuPeXI: prediction of neo-epitopes from tumor sequencing data. Cancer Immunol Immunother. 2017;66:1123–1130. doi: 10.1007/s00262-017-2001-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Castle JC, Loewer M, Boegel S, et al. Immunomic, genomic and transcriptomic characterization of CT26 colorectal carcinoma. BMC Genom. 2014;15:190. doi: 10.1186/1471-2164-15-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mosely SIS, Prime JE, Sainson RCA, et al. Rational selection of syngeneic preclinical tumor models for immunotherapeutic drug discovery. Cancer Immunol Res. 2017;5:29–41. doi: 10.1158/2326-6066.CIR-16-0114. [DOI] [PubMed] [Google Scholar]
- 4.DeVette CI, Andreatta M, Bardet W, et al. NetH2pan: a computational tool to guide MHC peptide prediction on murine tumors. Cancer Immunol Res Canimm. 2018 doi: 10.1158/2326-6066.CIR-17-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kreiter S, Vormehr M, van de Roemer N, et al. Mutant MHC class II epitopes drive therapeutic immune responses to cancer. Nature. 2015;520:692–696. doi: 10.1038/nature14426. [DOI] [PMC free article] [PubMed] [Google Scholar]