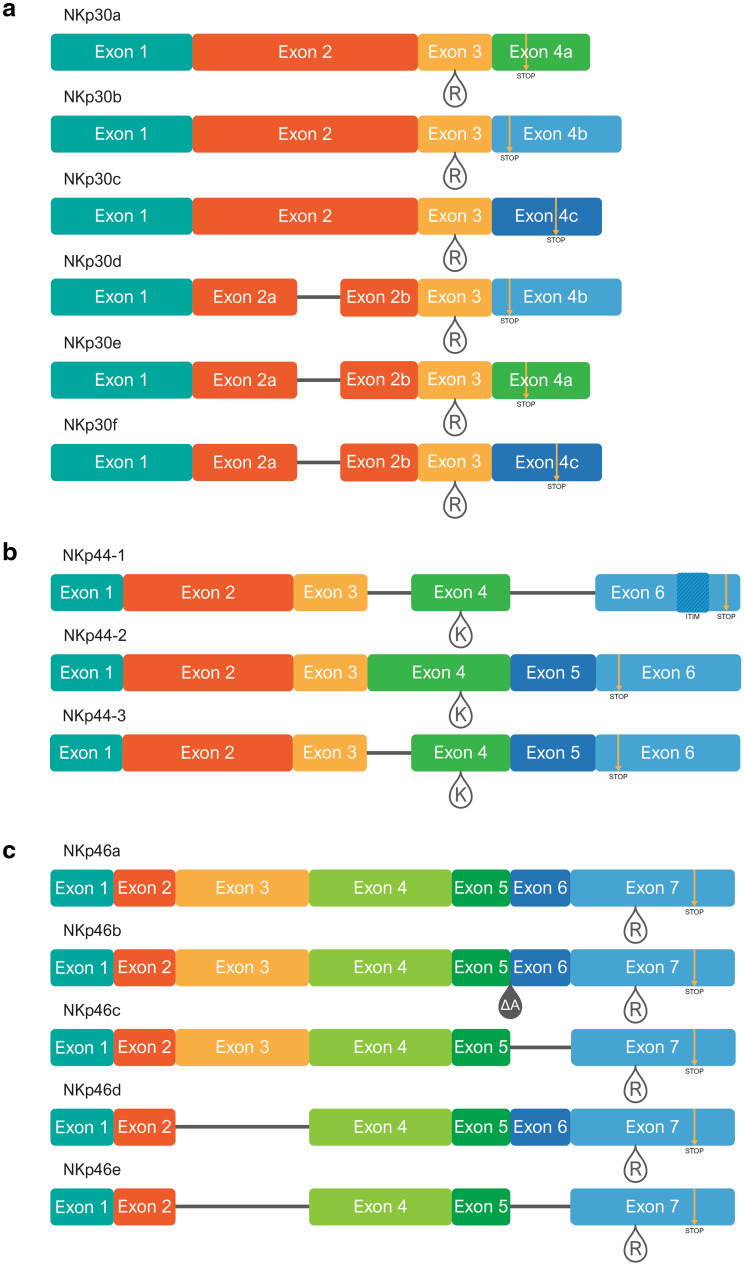
Fig. 2.
Exon map of NCR splice variants; a NKp30, b NKp44, and c NKp46. Regions representing the various exons are colored differently, and in accordance with the colors of the exon-encoded protein domains in Fig. 1. Teardrops mark locations of charged residues in the transmembrane domains (R = arginine, K = lysine). Yellow arrows mark premature stop codons caused by frame shifts during differential splicing. Location of the ITIM-like sequence is marked with hashed shading in NKp44-1. The ΔA teardrop in NKp46b marks the location of an alanine (A) deletion