Abstract
Immunotoxins are fusion proteins that combine a targeting component such as an antibody fragment or ligand with a cytotoxic effector component that induces apoptosis in specific cell populations displaying the corresponding antigen or receptor. Human cytolytic fusion proteins (hCFPs) are less immunogenic than conventional immunotoxins because they contain human pro-apoptotic enzymes as effectors. However, one drawback of hCFPs is that target cells can protect themselves by expressing endogenous inhibitor proteins. Inhibitor-resistant enzyme mutants that maintain their cytotoxic activity are therefore promising effector domain candidates. We recently developed potent variants of the human ribonuclease angiogenin (Ang) that were either more active than the wild-type enzyme or less susceptible to inhibition because of their lower affinity for the ribonuclease inhibitor RNH1. However, combining the mutations was unsuccessful because although the enzyme retained its higher activity, its susceptibility to RNH1 reverted to wild-type levels. We therefore used molecular dynamic simulations to determine, at the atomic level, why the affinity for RNH1 reverted, and we developed strategies based on the introduction of further mutations to once again reduce the affinity of Ang for RNH1 while retaining its enhanced activity. We were able to generate a novel Ang variant with remarkable in vitro cytotoxicity against HL-60 cells and pro-inflammatory macrophages. We also demonstrated the pro-apoptotic potential of Ang-based hCFPs on cells freshly isolated from leukemia patients.
Electronic supplementary material
The online version of this article (doi:10.1007/s00262-015-1763-8) contains supplementary material, which is available to authorized users.
Keywords: Angiogenin, RNH1, Human cytolytic fusion protein, Site-directed mutagenesis, Targeted therapy, Leukemia
Introduction
Human cytolytic fusion proteins (hCFPs) overcome the potential immunogenicity of immunotoxins while maintaining target cell specificity. Whereas conventional immunotoxins contain toxins derived from bacteria or plants, hCFPs are equipped with human enzymes as effector domains. Because these are fused with chimeric or fully human antibody fragments, the immunogenicity of hCFPs is expected to be minimal. However, the cytotoxicity of hCFPs is usually lower than that of conventional immunotoxins due to the presence of endogenous inhibitor proteins that mitigate the pro-apoptotic activity of human enzymes [1].
The eukaryotic ribonuclease inhibitor RNH1 binds and efficiently inactivates several members of the pancreatic ribonuclease superfamily, thereby reducing their enzymatic activity [2]. RNH1 is expressed in most mammalian cells and makes up ≥0.01 % of the total intracellular protein content [3]. Some non-human cytotoxic ribonucleases (e.g., ranpirnase/onconase and BS-RNase) are less sensitive or even insensitive to RNH1 [2]. The ability to evade RNH1 is therefore one of the major factors that determines ribonuclease cytotoxicity [4]. Inhibitor-sensitive hCFPs are only effective if the dose is sufficient to saturate the intracellular content of RNH1.
We recently developed variants of the human ribonuclease angiogenin (Ang) as improved hCFP effector domains [5]. Ang cleaves cellular tRNAs at the 3′ CCA terminus and at particular anticodon loops to generate tiRNAs [6]. These events lead to the shutdown of translation and promote the induction of apoptosis. Although the mechanistic basis of its pro-apoptotic activity remains unclear, Ang is fully suppressed by RNH1 under physiological conditions [7].
We modified Ang by site-directed mutagenesis to enhance its cytotoxicity and accordingly recovered one variant with lower affinity for the inhibitor (Ang GGRRmut) and another with higher enzymatic activity (Ang QGmut). However, when both mutations were combined in the same protein, the high affinity for RNH1 was restored and the mutant was no more potent than the wild-type enzyme. Molecular dynamic simulations revealed three interaction clusters in the RNH1-Ang complex [Cong et al. unpublished data]. Cluster I is conserved among all mutants and includes the N terminus of Ang. Cluster II contains the GGRR exchanges that promote the disassociation of Ang from RNH1. In the RNH1-Ang GGRRmut complex, cluster I has a hinge-like function that prevents the C terminus (cluster III) from interacting with the inhibitor and thus reduces the binding affinity. The QG exchange removes an obstruction covering the Ang active site by eliminating a hydrogen bond that stabilizes the C terminus in front of it [8, Cong et al. unpublished data]. This enhanced flexibility leads to an open enzyme conformation with improved substrate accessibility, but when combined with the GGRR mutation this C-terminal flexibility facilitates the recovery of cluster III and thus increases the affinity for RNH1.
One strategy to destabilize the interaction between RNH1 and Ang GGRR/QGmut while maintaining the enhanced enzyme activity conferred by the Q117G exchange is to weaken cluster I by introducing the exchanges R5A and H8A at sites known to participate in RNH1 binding [9, 10]. K40 also contributes to RNH1 binding because the K40Q exchange increases the K i by 1300-fold [9]. Although K40 is part of the catalytic triad, we generated the mutant Ang GGRR/QG/KQmut to determine whether minimal catalytic activity allows the induction of apoptosis combined with a low affinity for the inhibitor [11]. We also introduced the D116H exchange at the Ang C terminus because this increases the enzyme activity similarly to QG but replaces a negatively charged amino acid with a positively charged one [12] which should prevent the recovery of cluster III.
Here, we report the functionality of these novel angiogenin mutants fused with the CD64-specific single-chain variable fragment (scFv) H22 [13, 14]. The fusion proteins were transiently expressed in HEK293T cells and characterized in terms of target cell binding, enzyme activity, susceptibility to inhibition and cytotoxic potency. Human pro-inflammatory macrophages (hM1Φ) and the promyelocytic cell line HL-60 were used as target cells in vitro. Peripheral blood mononuclear cells (PBMCs) were isolated from acute or chronic myeloid leukemia (AML/CMML) patients and were used as target cells ex vivo.
Materials and methods
Site-directed mutagenesis
The plasmid containing the H22-Ang GGRR/QGmut sequence was modified using splicing by overlap extension PCR (SOE-PCR) with Taq Phusion DNA polymerase (New England Biolabs, Ipswich, MA, USA) as previously described [5, 15]. Exchange oligonucleotide primers were designed and synthesized by Eurofins Genomics (Ebersberg, Germany) (Supplementary Table 1). The first two DNA fragments were amplified using 0.5 µM of each mutagenesis primer by heating to 98 °C for 1 min followed by 30 cycles of 98 °C for 50 s, 73 °C for 50 s and 72 °C for 40 s, with a final extension step at 72 °C for 2 min. The fragments were separated by 1.2 % agarose gel electrophoresis and purified using the NucleoSpin Gel and PCR Clean-up kit (Macherey & Nagel, Düren, Germany). The second PCR was carried out using 10 ng of each fragment for end annealing and overlap extension. The reaction was heated to 95 °C for 5 min followed by eight cycles of 95 °C for 45 s, 55 °C for 45 s and 72 °C for 1 min, with a final extension step at 72 °C for 7 min. We then added 0.2 µM of the 5′-SfiI_for and Hinterhis_rev primers to 10 ng of the self-annealed and extended fragment, and heated the reaction to 95 °C for 2 min followed by 30 cycles of 95 °C for 45 s, 55 °C for 45 s and 72 °C for 1 min, with a final extension step at 72 °C for 10 min. After fragment purification and digestion with XbaI and BlpI (New England Biolabs), the DNA fragment was inserted into the pMS expression vector [16]. Base exchanges were verified by DNA sequencing using an ABI Prism 3700 DNA Analyzer and BigDye cycle sequencing terminator chemistry (Applied Biosystems, Waltham, MA, USA).
Cell culture, hCFP expression and purification
HL-60 (ATCC, CCL-240) and L-540cy cells (kindly provided by Prof. Dr. med. Andreas Engert, University Hospital Cologne, Germany) were cultivated in RPMI 1640 medium with GlutaMAX™, 10 % fetal calf serum and 5 % penicillin/streptomycin (Life Technologies, Darmstadt, Germany) under standard conditions (37 °C, 5 % CO2) without selection [17, 18]. Cell cultivation, transient hCFP expression, protein purification by immobilized metal ion affinity chromatography (IMAC) and the determination of hCFP concentrations were carried out as previously described [5].
Preparation of hMΦ cells from buffy coat-derived PBMCs
Buffy coats were obtained from the Department of Transfusion Medicine, University Hospital RWTH Aachen, Germany. PBMCs were isolated and polarized to produce a population of hM1Φ cells as previously described [19]. For subsequent analysis, cells were seeded into 96-well cell culture plates at a density of 5 × 105 cells per ml.
Preparation of PBMCs from blood and bone marrow specimens
All leukemia specimens were kindly provided by the Department of Hematology and Oncology (Internal Medicine IV, University Hospital RWTH Aachen) after informed consent with the approval of the clinical research ethics board of the RWTH Aachen and pre-screening for CD64 expression. None of the patients received any form of therapy before sample donation. Bone marrow specimens were filtered using 100 µm pluriStrainer filter units (pluriSelect, San Diego, CA, USA) before diluting 1:5 with sterile DPBS (Life Technologies). Peripheral blood was diluted 1:3 in DPBS without filtration. PBMCs were isolated by density centrifugation as previously described [19], and 8 × 105 cells per ml were cultivated in RPMI 1640 medium with 10 % heat-inactivated human serum (Sigma-Aldrich, Steinheim, Germany) in the presence of 100 U/ml human interferon gamma (hIFNγ, Sigma) at 37 °C and 5 % CO2.
Flow cytometry
The analysis of hCFP binding to cultured cells was carried out using a BD FACSVerse flow cytometer (Becton–Dickinson, Heidelberg, Germany) and BD FACSuite software according to the manufacturer’s instructions. We stimulated 5 × 105 HL-60 cells per sample with 300 U/ml hIFNγ 24 h before analysis, with L-540cy cells as negative controls. Polarized hM1Φ cells were prepared as described above. The cells were incubated on ice with 1 µg of the purified hCFPs for 30 min before adding 1 µg/ml anti-His5 Alexa Fluor 488 conjugate (Qiagen, Hilden, Germany) detection antibody and incubating for 20 min in the dark. The cells were washed between incubation steps with cold 1x PBS. We analyzed 104 HL-60 and L-540cy cells in each sample, or the entire population of PBMCs. For the latter, the hCFPs were supplemented with 1 % heat-inactivated human serum to increase binding specificity.
In vitro tRNA cleavage assays
The tRNA cleavage assays were carried out as previously described [5]. Substrate cleavage in the presence of the inhibitor was demonstrated by adding 5 U Ribolock RNase inhibitor (Life Technologies) to 1 µg hCFP under the same cleavage conditions.
Quantitative analysis of ribonucleolytic activity and inhibition
We incubated 5 pmol (50 nM) RNaseAlert™ (Integrated DNA Technologies, Leuven, Belgium) with 10 pmol (100 nM) hCFP overnight at room temperature in RNase-free cleavage buffer (30 mM Tris/NaCl, pH 8.0) in a total reaction volume of 100 µl per well. The RNaseAlert™ substrate is a fluorophore- and quencher-coupled RNA sequence optimized for ribonucleolytic digestion. When exposed to the excitation wavelength (485 nm), only cleaved substrates emit a signal (520 nm) because the signal from the intact substrate is quenched. All samples were tested at least in duplicate in black µClear 96-well plates (655090, Greiner Bio-One, Frickenhausen). H22(scFv)-SNAP was used as negative control fusion protein under the same conditions to demonstrate cleavage specificity. The substrate dissolved in reaction buffer was used to correct for background fluorescence [20]. Fluorescence intensities were measured using a Tecan Genios Pro microplate reader (Tecan, Mainz, Germany) with Magellan v7.1 SP1 software. Measurements were taken 18 times at 3-min intervals to ensure the cleavage reaction was completed. The values were averaged and converted into relative enzyme activities by signal normalization against the background fluorescence (0 %) and the H22-Ang wild-type fluorescence signal (100 %). Inhibition was measured by supplementing the reaction with decreasing quantities of RiboLock™ (0.02, 0.015, 0.005, 0.0016, 0.00056 and 0.00019 nmol, corresponding to 200–2 nM). The relative enzymatic activities at each RNH1 concentration were calculated by signal normalization against background fluorescence (0 %) and the individual maximum fluorescence of each hCFP without inhibition (100 %). The logarithmic inhibitor quantity was plotted against the relative enzyme activities, and the sigmoidal dose–response curve was fitted using GraphPad Prism v5 with a log (inhibitor) versus response algorithm.
XTT proliferation assay
XTT assays were carried out as previously described on HL-60, L-540cy [19] and hM1Φ cells [5]. Raw data were evaluated using Microsoft Excel 2010 and GraphPad Prism v5. Cytotoxicity was calculated from a sigmoidal dose–response fit with a variable slope.
Apoptosis assays on leukemia cells
PBMCs isolated from leukemia patients were seeded as described above and incubated with 200 nM hCFPs or controls for 12 h with an end volume of 600 µl per well. Negative control cells were untreated, and 30 µl camptothecin was added as an internal positive control. Specific pro-apoptotic effects were confirmed by applying irrelevant hCFPs containing 2112(scFv), anti-her2(scFv) and myelin oligodendrocyte glycoprotein (MOG)-based binding components under equivalent conditions [21, 22, Bialon et al. unpublished data]. Truncated Pseudomonas aeruginosa exotoxin A (ETA′) fused with H22 was used as a reference immunotoxin [23]. Each sample was tested at least in duplicate. The cells were then scraped and washed with 1x annexin V binding buffer (10 mM HEPES, 140 mM NaCl, 2.5 mM CaCl2, pH 7.4). Following centrifugation (1500 rpm, 7 min, 4 °C), the pellet was resuspended in 450 µl annexin V-eGFP saturated cell culture supernatant and 50 µl 10× annexin V binding buffer [24]. After incubation for 15 min in the dark, the cells were washed with 1× PBS, resuspended in 1× annexin V binding buffer and supplemented with 10 µg/ml propidium iodide (PI) before measurement. Fluorescence compensation was carried out with camptothecin-treated cells stained with either annexin V-eGFP or PI. The induction of apoptosis was confirmed using flow cytometry by counting 104 cells in total and plotting fluorescence channel FL-1 against FL-3 [25]. The sum of annexin V-positive cells (early-apoptotic and late-apoptotic cells) was calculated for the control and hCFP-treated samples, respectively.
Statistical analysis
Statistical significance was determined by applying a two-tailed Student’s t test using GraphPad Prism v5. Values are expressed as means ± standard deviations (SD) or standard errors of the mean (SEM) as indicated (*p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001).
Results
Construction, expression and purification of mutant hCFPs
The H22-Ang variants GGRR/QG/RAmut, GGRR/QG/HAmut, GGRR/QG/DHmut and GGRR/QG/KQmut were prepared by site-directed mutagenesis and verified by DNA sequencing. After transient expression in HEK293T cells, the hCFPs were purified by IMAC. The yields ranged from 0.1 to 0.9 mg per liter of cell culture supernatant, and the purity ranged from 60 to 90 %, as estimated by staining with Coomassie Brilliant Blue followed by quantification with “advanced image data analyzer” (AIDA) software.
Cleavage of yeast tRNA in vitro by recombinant hCFPs
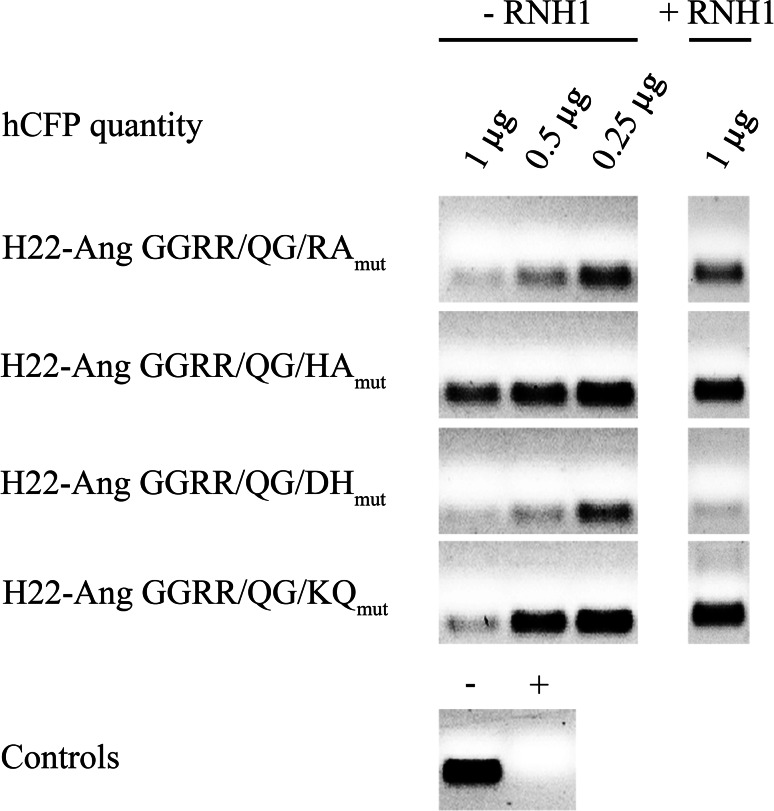
The in vitro cleavage of tRNA by each hCFP was tested in the presence and absence of the inhibitor (Fig. 1). RNase A was used as a positive control, and yeast tRNA in sample buffer was used as a negative control. Different concentrations (0.25–1 µg) of each hCFP were tested, and all four proteins showed dose-dependent ribonucleolytic activity. H22-Ang GGRR/QG/DHmut displayed similar activity in the presence and absence of RNH1, whereas the others were inhibited by RNH1 to different degrees, with H22-Ang GGRR/QG/KQmut appearing most susceptible to inhibition.
Fig. 1.
Representative yeast tRNA cleavage assay. The hCFP-mediated degradation of tRNA was demonstrated by incubating 1–0.25 µg of each fusion protein with 600 ng yeast tRNA for 90 min at 37 °C to measure the dose-dependent degradation efficiency. We used 100 ng RNase A as a positive control (+) and tRNA in RNase-free reaction buffer as a negative control (−). The degradation efficiency in the presence of RNH1 was tested in a representative experiment by adding 5 U commercial RNH1 to 1 µg hCFP under the same cleavage conditions. H22-Ang GGRR/QG/DHmut achieved the highest cleavage efficiency in the presence of RNH1
Quantitative analysis of hCFP ribonucleolytic activity
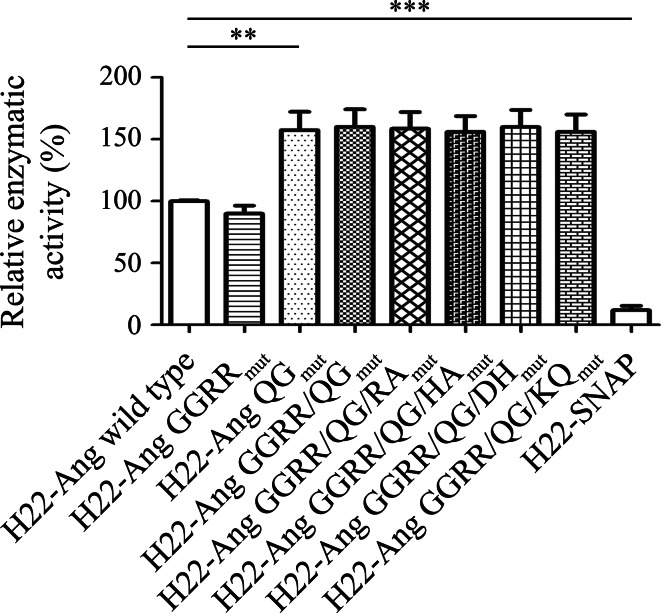
The ribonucleolytic activity of the four new hCFPs (100 nM) was compared to the previously described variants H22-Ang GGRRmut, H22-Ang QGmut and H22-Ang GGRR/QGmut [5], and the wild-type enzyme, using RNaseAlert™ with H22-SNAP as a negative control. The fluorescence signal for wild-type H22-Ang after overnight incubation was set to 100 %, and the background fluorescence was set to 0 % to provide reference values for the remaining constructs (Fig. 2). The activity of all four new variants was similar to that of H22-Ang QGmut and H22-Ang GGRR/QGmut, and was superior to that of the wild-type enzyme and variant H22-Ang GGRRmut as previously reported [5].
Fig. 2.
Relative enzymatic activities of hCFPs. The hCFPs were incubated with RNaseAlert™ overnight at room temperature, and the fluorescence emission was normalized against the fluorescence background (0 %) and the maximal fluorescence of wild-type H22-Ang (100 %). H22-SNAP was used as a negative control under the same conditions. Relative enzymatic activities are expressed as means of three independent experiments ± SEM. Statistical significance was determined using a two-tailed unpaired Student’s t test (*p ≤ 0.05, **p ≤ 0.01 and ***p ≤ 0.001)
Susceptibility of the hCFPs to inhibition by RNH1
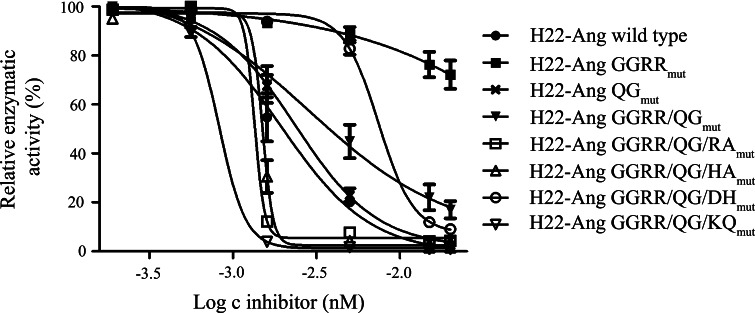
Individual inhibition profiles were prepared for all hCFPs by comparing their relative activities in the presence of different concentrations (2–200 nM) of the inhibitor (Fig. 3). Wild-type H22-Ang, H22-Ang GGRRmut and H22-Ang GGRR/QGmut were used as reference proteins [5]. Following hCFP-mediated RNaseAlert™ cleavage as described above, the fluorescence intensities were converted to relative activities by setting the fluorescence background to 0 % and the individual maximum fluorescence intensities without inhibition to 100 %. Sigmoidal and dose-dependent inhibition profiles were generated after plotting the relative activities against the concentrations of the inhibitor. As previously indicated [5], H22-Ang QGmut and H22-Ang GGRR/QGmut were inhibited to the same degree as wild-type H22-Ang, whereas H22-Ang GGRRmut retained its full activity at all inhibitor concentrations. Among the four new constructs, H22-Ang GGRR/QG/RAmut, H22-Ang GGRR/QG/HAmut and H22-Ang GGRR/QG/KQmut appeared to be more susceptible to the inhibitor than the wild-type protein, whereas H22-Ang GGRR/QG/DHmut was more resistant and only lost activity at the highest tested inhibitor concentration.
Fig. 3.
Susceptibility of hCFPs to inhibition by RNH1. We incubated 100 nM of each hCFP with 50 nM of RNaseAlert™ and 2–200 nM RNH1 overnight at room temperature, and the fluorescence emission was normalized against the fluorescence background (0 %) and the desired maximum fluorescence intensities without inhibition (100 %). Individual inhibition profiles were obtained by plotting the logarithmic protein concentration of RNH1 against the relative enzymatic activities. The sigmoidal dose-dependent inhibition was fitted using a log (inhibitor) versus response fit. Values are expressed as mean ± SEM of three independent experiments
In vitro binding of hCFPs and depletion of stimulated HL-60 cells and hM1Φ cells
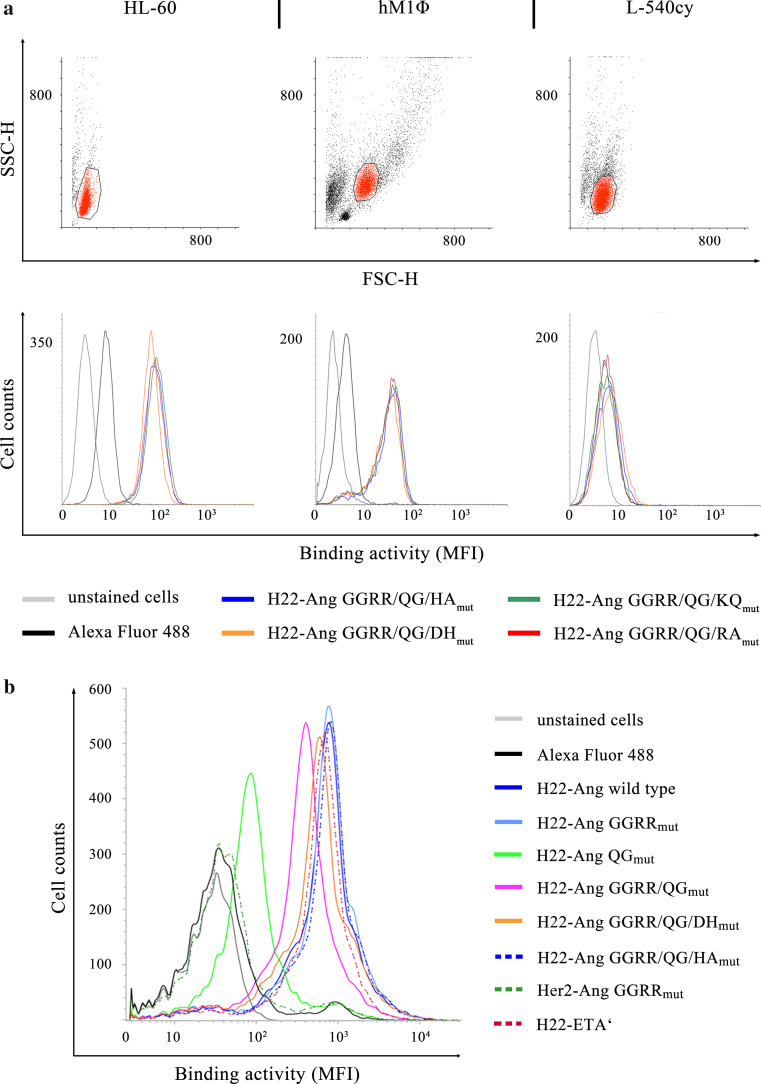
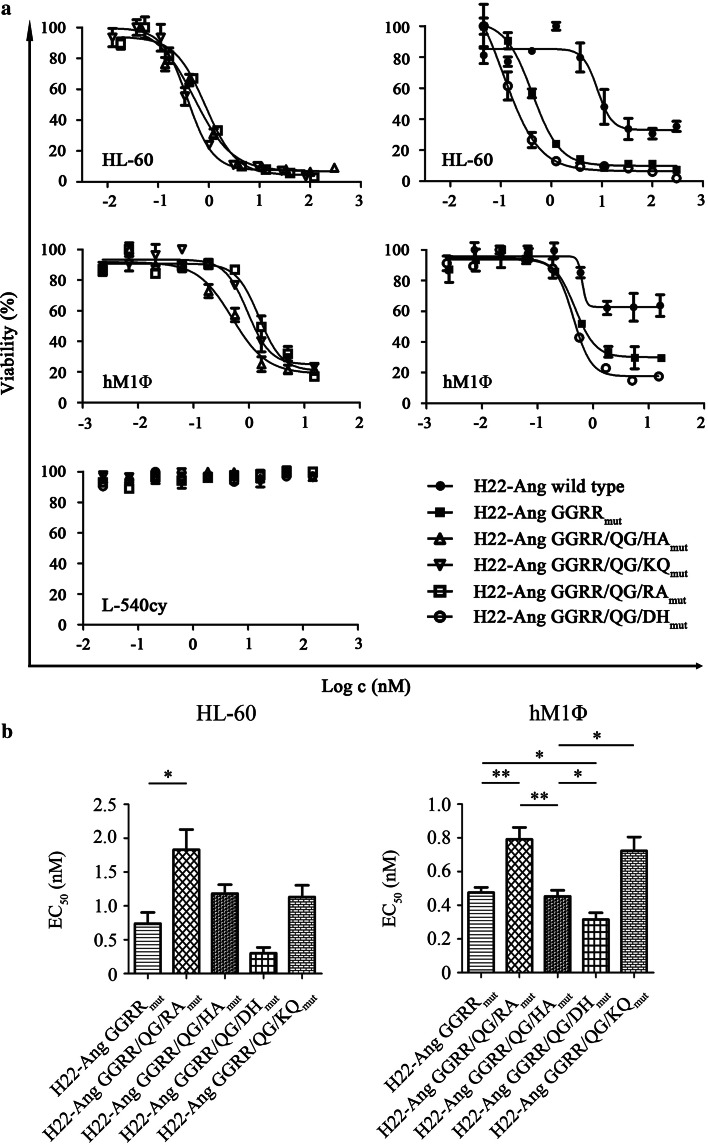
The cell-specific binding activity of all hCFPs was confirmed by flow cytometry using stimulated HL-60 cells, hM1Φ cells and leukemia-derived PBMCs. No binding to CD64− L-540cy cells was observed (Fig. 4a, b). The cytotoxicity of each hCFP against stimulated HL-60 cells and human M1Φ cells was subsequently demonstrated using cell viability assays. Serial 1:3 dilutions were applied to the cells starting at 300 nM and 15 nM, respectively. Viable cells were quantified using an XTT assay after exposure to the hCFPs for 72 h (Fig. 5a). The specific cytotoxicity of each hCFP was also verified by the absence of any effect against the CD64− cell line L-540cy. The corresponding EC50 values are presented in Supplementary Table 2. Using H22-Ang GGRRmut as reference for cytotoxicity, the statistical comparison of EC50 values revealed that H22-Ang GGRR/QG/DHmut showed slightly (but significantly) greater toxicity toward hM1Φ cells and also slightly (but not significantly) greater toxicity toward HL-60 cells (Fig. 5b). Compared to H22-Ang GGRRmut, the remaining hCFPs showed similar or lower levels of cytotoxicity toward both cell types. Compared to wild-type H22-Ang, H22-Ang GGRRmut was up to 12-fold more cytotoxic toward HL-60 cells, whereas H22-Ang GGRR/QG/DHmut was at least 13-fold more cytotoxic toward HL-60 cells.
Fig. 4.
In vitro and ex vivo binding activities of hCFPs. We incubated 1 µg hCFP with hIFNγ-stimulated HL-60, hM1Φ and L-540cy cells (a) or leukemia-derived PBMCs (b) for 30 min on ice. An Alexa Fluor 488 detection antibody was added to the samples for 20 min on ice in the dark, and binding was demonstrated by flow cytometry. A fluorescence shift compared to the detection antibody control indicated binding. The HL-60, L–540cy and hM1Φ target cell populations were gated as indicated (a, SSC-H/FSC-H), whereas all PBMCs were analyzed for specific binding
Fig. 5.
Cytotoxicity of hCFPs against HL-60 and hM1Φ cells. Stimulated HL-60 and hM1Φ cells (a) were used to evaluate hCFP cytotoxicity. The cells were exposed to serial dilutions of hCFPs for 72 h and treated with either zeocin or PBS as controls. Viable cells were detected using a XTT colorimetric assay. Target cell specificity was demonstrated by the absence of cytotoxicity against CD64− L-540cy cells. EC50 values were calculated after sigmoidal dose–response fitting (b). EC50 values are expressed as mean ± SD from at least three independent experiments. Statistical comparisons were carried out using a two-tailed unpaired Student’s t test (*p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001)
Reduction in leukemia cell viability ex vivo by hCFPs
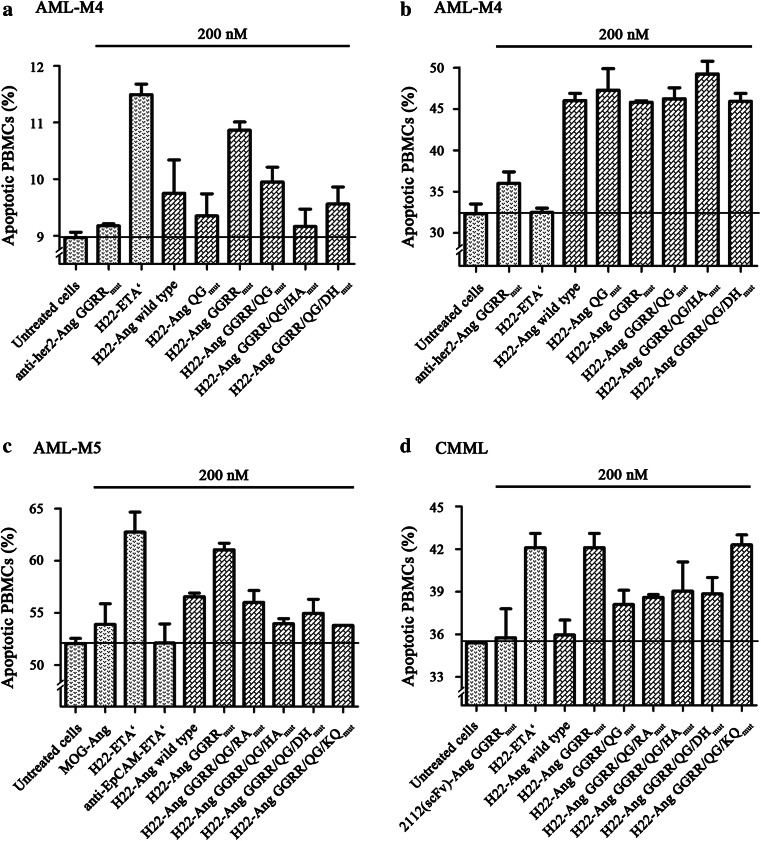
The pro-apoptotic effect of each hCFP was tested against PBMCs from untreated leukemia patients with varying subtypes (Fig. 6). Clinically relevant patient data are summarized in Supplementary Table 3. For the apoptosis assays, 8 × 105 PBMCs/ml were cultivated in the presence of human serum containing 100 U/ml hIFNγ. We then added 200 nM of each hCFP and incubated the cells for 12 h before counting the apoptotic cells by flow cytometry. The sum of early-apoptotic and late-apoptotic cells was compared to appropriate controls (Fig. 6). No reduction in cell viability compared to the medium-only control was observed following the addition of 2112-Ang GGRRmut, anti-her2-Ang GGRRmut or MOG-Ang (Fig. 6, samples a–d). H22-ETA′ induced apoptosis in samples a, c and d but not b. However, all the hCFPs reduced cell viability in sample b. Similar pro-apoptotic effects were observed for H22-ETA′ and H22-Ang GGRRmut, whereas H22-Ang GGRR/QG/KQmut was cytotoxic toward samples b and d but not the others. The remaining hCFPs showed similar pro-apoptotic activity against all four samples, although the potency varied.
Fig. 6.
Representative ex vivo depletion of isolated leukemia PBMCs. PBMCs were isolated by density gradient centrifugation from the peripheral blood (a–c) or bone marrow (d) of acute or chronic myelomonocytic leukemia patients. The cells were cultivated in the presence of 200 nM hCFPs and controls for 12 h at 37 °C in a 5 % CO2 atmosphere. The cells were then double-stained with annexin V-eGFP and PI before analysis by flow cytometry. The sum of early-apoptotic and late-apoptotic cells is shown as the mean of duplicates ± SD in comparison with the controls
Discussion
The aim of this study was to optimize our previously reported Ang variants by introducing four additional point mutations (R5A, H8A, K40Q and D116H) into the Ang GGRR/QGmut variant. The novel variants were then joined to the scFv H22 for evaluation [5]. The hCFPs were transiently expressed in HEK293T cells, and the yields were similar to those achieved for H22-Ang GGRRmut, Ki4-Ang, Ang-Ki4 [5, 16], Ang-MJ7, MJ7-Ang and RFB4-Ang [26, 27]. We observed a general inverse correlation between the yield and cytotoxicity of each hCFP, suggesting comparative expression studies may be useful to identify the most suitable expression system.
Each of the hCFPs showed a similar and highly specific in vitro binding activity. The observed differences in preference for leukemia-derived PBMCs probably reflected varying CD64 expression levels based on donor specificities and leukemia subtypes. The inclusion of human serum was anticipated to reduce nonspecific binding events [28]. However, these stringent conditions may have reduced the binding efficiency, thus contributing to the smaller fluorescence shifts.
The catalytic activity of each hCFP was tested by tRNA cleavage in vitro. Although this assay showed that the different constructs varied in their cleavage efficiency, precise quantification was challenging because it depends on individual densitometric measurements. Quantitative analysis was therefore carried out using the RNaseAlert™ substrate. The activity of H22-Ang GGRRmut was similar to that of wild-type H22-Ang, but it was outperformed by H22-Ang QGmut, as previously reported [5]. All variants containing the Q117G exchange showed similar activity. Interestingly, the K40Q exchange did not affect the activity of the enzyme in this assay despite previous reports that the active center was disrupted and the ribonucleolytic activity was reduced by 2 × 103-fold compared to the wild type [29]. The prolonged cleavage time for RNaseAlert™ compared to tRNA may have allowed the digestion of similar amounts of substrate, despite differences in catalytic activity (Figs. 1, 2). At a certain point, unfavorable cleavage conditions prevent further digestion. Differential preferences for tRNA and the artificial substrate may also explain our observations.
We anticipated a weaker reduction in the individual relative enzymatic activities of variants with a lower affinity for RNH1 and used the individual signal deprivation to quantify the degree of inhibition. We found that H22-Ang QGmut, H22-Ang GGRR/QGmut and H22-Ang wild-type were similarly susceptible to inhibition, confirming our previous data and molecular dynamic simulations [5, Cong et al. unpublished data]. Ang GGRRmut has a 106-fold lower affinity for RNH1 as shown by its potent activity even at high RNH1 concentrations (Fig. 3) [30]. Mutations R5A and H8A are close together and reduce the affinity of wild-type Ang for RNH1 by 50-fold and 4.6-fold, respectively [9, 10]. These mutations were introduced to weaken cluster I, while GGRR mediates the disengagement of cluster II from RNH1. However, both variants appeared to be more susceptible to RNH1 than wild-type H22-Ang. R5A and H8A may weaken but not fully disrupt cluster I, which would allow both variants to eradicate the steric incompatibility caused by GGRR. Furthermore, whereas the disassociation at cluster II is retained, the relaxed cluster I interaction potentially promotes the formation of tighter interactions at cluster III. Although an extraordinary increase in Ki was reported for Ang K40Q, H22-Ang GGRR/QG/KQmut demonstrated the greatest susceptibility to RNH1 indicating that it is difficult to predict the consequences of multiple simultaneous enzyme modifications. H22-Ang GGRR/QG/DHmut retained its full enzymatic activity until exposed to the highest inhibitor concentration and featured a more potent ribonucleolytic activity than wild-type H22-Ang.
The cytotoxic potency of wild-type H22-Ang was similar to that of granzyme M-H22 against HL-60 cells or granzyme B-H22 against U937 cells [31, 32]. Most of the novel hCFPs were approximately fourfold more cytotoxic against HL-60 cells than the wild-type H22-Ang but were less cytotoxic than H22-Ang GGRRmut. The exception was H22-Ang GGRR/QG/DHmut, which outperformed H22-Ang GGRRmut due to its higher ribonucleolytic activity in the presence of moderate-to-high concentrations of the inhibitor. The EC50 of H22-Ang GGRR/QG/DHmut was similar to that of H22-ETA′ against U937 cells, demonstrating the potential of optimized hCFPs to compete with traditional immunotoxins [23]. H22-Ang GGRRmut and H22-Ang GGRR/QG/DHmut also showed the most potent activity against hM1Φ cells, although the cytotoxicity of H22-Ang GGRRmut was lower than previously described [5]. This may reflect the heterogeneity of cell sensitivity depending on donor properties, the RNH1 content and the efficiency of cell stimulation.
The leukemia cells used for ex vivo depletion studies represented different subtypes (e.g., AML and CMML) with varying frequencies of CD64+ cells and CD64 expression levels [33, 34]. The small sample number, specimen heterogeneity and inability to reproduce patient-specific assays did not facilitate statistical analysis. The addition of hIFNγ promotes monocytic CD64 expression and was previously shown to improve the sensitivity of H22-based cell targeting [31, 32, 35, 36]. CD64 expression and hCFP binding activity both affect the cytotoxic potency of hCFPs by limiting ribonuclease delivery. However, this does not explain the varying efficacy of the hCFPs against individual specimens. Only H22-Ang GGRRmut demonstrated reproducible pro-apoptotic effects. Similar observations were reported for the inhibitor-insensitive hCFP GbR201K-H22(scFv), suggesting that lower inhibitor affinity is a key determinant of cytotoxicity [35]. A prolonged incubation time might improve the potency of the remaining hCFPs, but we limited the assay to 12 h because some cell samples were sensitive toward treatment-independent apoptosis. The pro-apoptotic effects of the new hCFPs must therefore be compared to H22-ETA′ as a reference. H22-Ang GGRRmut was at least as cytotoxic as H22-ETA′ and was effective against cells that are unaffected by this immunotoxin (Fig. 6b), confirming the results previously reported for GbR201K-H22(scFv). ETA′ resistance was first observed in children with acute lymphoblastic leukemia and is caused by a deletion in the WDR85 gene [37]. However, these cells are not protected against hCFP treatment, which highlights the promising aspects of human enzymes. The observed ex vivo cytotoxicity of the remaining variants did not correlate with the corresponding in vitro data. Leukemia types and even subtypes are characterized by substantial genetic heterogeneity [38, 39] involving signaling pathways that regulate the cell cycle, proliferation, survival and apoptosis [40, 41]. Ang participates directly or indirectly in diverse signaling pathways, including ERK1/2, B/akt SAPK/JNK and PI3K/AKT/mTOR, among which at least the former can promote apoptosis [42–44]. Ang also co-immunoprecipitates with p53 and Mdm2 and may affect their regulatory functions [45]. These observations suggest that Ang promotes translational shutdown to protect cells from stress [46]. Apoptosis may be promoted by multiple processes, including proliferation arrest and pro-apoptotic signaling through pathways that might be altered or deregulated, with different effects on individual Ang mutants as effector domains.
In summary, we generated the novel Ang variant GGRR/QG/DHmut and showed that it is up to 30-fold more cytotoxic than wild-type H22-Ang against HL-60 cells. The improvement reflects its higher ribonucleolytic activity combined with greater resistance against the inhibitor RNH1. H22-Ang GGRRmut shows reproducible pro-apoptotic activity against CD64+ leukemia cells ex vivo, with a similar efficacy to the reference protein H22-ETA′. The protein was also cytotoxic toward cells that are resistant to H22-ETA′. Our results suggest that hCFPs with improved properties have the potential to compete with or even outperform traditional immunotoxins and that in silico simulations can facilitate the development of mutant variants with optimized characteristics.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This project was funded by the Deutsche Forschungsgemeinschaft (DFG). The authors would like to thank Dr. Christoph Stein (Institute for Applied Medical Engineering, University Hospital RWTH Aachen/Fraunhofer Institute for Molecular Biology and Applied Ecology, Department of Pharmaceutical Product Development, Aachen) for helpful discussions on leukemia specimen handling, Judith Niesen (Fraunhofer Institute for Molecular Biology and Applied Ecology, Department of Pharmaceutical Product Development, Aachen) for providing the fusion protein 2112(scFv)-Ang GGRRmut, Anh-Tuah Pham (Institute for Applied Medical Engineering, University Hospital RWTH Aachen) for providing her2(scFv)-Ang GGRRmut and Fanny Frenzel (University Hospital RWTH Aachen, Department of Hematology and Oncology, Internal Medicine IV, Aachen, Germany) for providing patient data and specimens. Finally, we are very grateful to Dr. Richard M Twyman for critically reading this manuscript. Radoslav Mladenov was supported by a scholarship from the Jürgen Manchot Foundation.
Abbreviations
- AML
Acute myeloid leukemia
- Ang
Angiogenin
- CMML
Chronic myelomonocytic leukemia
- DNA
Deoxyribonucleic acid
- EC50
Half maximal effective concentration
- Gb
Granzyme B
- GFP
Green fluorescent protein
- hCFP
Human cytolytic fusion protein
- hIFNγ
Human interferon gamma
- hM1Φ
Human pro-inflammatory macrophages
- HEK293T
Human embryonic kidney cells
- IMAC
Immobilized metal ion affinity chromatography
- Ki
Inhibitory constant
- MOG
Myelin oligodendrocyte glycoprotein
- PBMC
Peripheral blood mononuclear cell
- PCR
Polymerase chain reaction
- PI
Propidium iodide
- RNA
Ribonucleic acid
- RNH1
Ribonuclease/angiogenin inhibitor 1
- RPMI
Roswell Park Memorial Institute
- SEM
Standard error of the mean
- SOE
Splicing by overlap extension
- tRNA
Transfer RNA
- tiRNA
tRNA-derived stress-induced RNA
- XTT
2,3-bis-(2-Methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide
Compliance with ethical standards
Conflict of interest
The authors have no conflict of interest to disclose.
Footnotes
Stefan Barth and Thomas Nachreiner contributed equally to this manuscript.
References
- 1.Weidle UH, Georges G, Brinkmann U. Fully human targeted cytotoxic fusion proteins: new anticancer agents on the horizon. Cancer Genomics Proteomics. 2012;9:119–133. [PubMed] [Google Scholar]
- 2.Monti DM, D’Alessio G. Cytosolic RNase inhibitor only affects RNases with intrinsic cytotoxicity. J Biol Chem. 2004;279:39195–39198. doi: 10.1074/jbc.C400311200. [DOI] [PubMed] [Google Scholar]
- 3.Leland PA, Raines RT. Cancer chemotherapy–ribonucleases to the rescue. Chem Biol. 2001;8:405–413. doi: 10.1016/S1074-5521(01)00030-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rutkoski TJ, Raines RT. Evasion of ribonuclease inhibitor as a determinant of ribonuclease cytotoxicity. Curr Pharm Biotechnol. 2008;9:185–189. doi: 10.2174/138920108784567344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cremer C, Vierbuchen T, Hein L, et al. Angiogenin mutants as novel effector molecules for the generation of fusion proteins with increased cytotoxic potential. J Immunother. 2015;38:85–95. doi: 10.1097/CJI.0000000000000053. [DOI] [PubMed] [Google Scholar]
- 6.Czech A, Wende S, Morl M, et al. Reversible and rapid transfer-RNA deactivation as a mechanism of translational repression in stress. PLoS Genet. 2013;9:e1003767. doi: 10.1371/journal.pgen.1003767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pizzo E, Sarcinelli C, Sheng J, et al. Ribonuclease/angiogenin inhibitor 1 regulates stress-induced subcellular localization of angiogenin to control growth and survival. J Cell Sci. 2013;126:4308–4319. doi: 10.1242/jcs.134551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Russo N, Shapiro R, Acharya KR, et al. Role of glutamine-117 in the ribonucleolytic activity of human angiogenin. Proc Natl Acad Sci USA. 1994;91:2920–2924. doi: 10.1073/pnas.91.8.2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen CZ, Shapiro R. Superadditive and subadditive effects of “hot spot” mutations within the interfaces of placental ribonuclease inhibitor with angiogenin and ribonuclease A. Biochemistry. 1999;38:9273–9285. doi: 10.1021/bi990762a. [DOI] [PubMed] [Google Scholar]
- 10.Chen CZ, Shapiro R. Site-specific mutagenesis reveals differences in the structural bases for tight binding of RNase inhibitor to angiogenin and RNase A. Proc Natl Acad Sci USA. 1997;94:1761–1766. doi: 10.1073/pnas.94.5.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Acharya KR, Shapiro R, Allen SC, et al. Crystal structure of human angiogenin reveals the structural basis for its functional divergence from ribonuclease. Proc Natl Acad Sci USA. 1994;91:2915–2919. doi: 10.1073/pnas.91.8.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leonidas DD, Shapiro R, Subbarao GV, et al. Crystallographic studies on the role of the C-terminal segment of human angiogenin in defining enzymatic potency. Biochemistry. 2002;41:2552–2562. doi: 10.1021/bi015768q. [DOI] [PubMed] [Google Scholar]
- 13.de Kruif J, Tijmensen M, Goldsein J, et al. Recombinant lipid-tagged antibody fragments as functional cell-surface receptors. Nat Med. 2000;6:223–227. doi: 10.1038/72339. [DOI] [PubMed] [Google Scholar]
- 14.Graziano RF, Tempest PR, White P, et al. Construction and characterization of a humanized anti-gamma-Ig receptor type I (Fc gamma RI) monoclonal antibody. J Immunol. 1995;155:4996–5002. [PubMed] [Google Scholar]
- 15.Ho SN, Hunt HD, Horton RM, et al. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 16.Stocker M, Tur MK, Sasse S, et al. Secretion of functional anti-CD30-angiogenin immunotoxins into the supernatant of transfected 293T-cells. Protein Expr Purif. 2003;28:211–219. doi: 10.1016/S1046-5928(02)00709-X. [DOI] [PubMed] [Google Scholar]
- 17.Gallagher R, Collins S, Trujillo J, et al. Characterization of the continuous, differentiating myeloid cell line (HL-60) from a patient with acute promyelocytic leukemia. Blood. 1979;54:713–733. [PubMed] [Google Scholar]
- 18.Diehl V, Kirchner HH, Schaadt M, et al. Hodgkin’s disease: establishment and characterization of four in vitro cell lies. J Cancer Res Clin Oncol. 1981;101:111–124. doi: 10.1007/BF00405072. [DOI] [PubMed] [Google Scholar]
- 19.Schiffer S, Hristodorov D, Mladenov R, et al. Species-dependent functionality of the human cytolytic fusion proteins granzyme B-H22(scFv) and H22(scFv)-angiogenin in macrophages. Antibodies. 2013;2:9–18. doi: 10.3390/antib2010009. [DOI] [Google Scholar]
- 20.Kampmeier F, Ribbert M, Nachreiner T, et al. Site-specific, covalent labeling of recombinant antibody fragments via fusion to an engineered version of 6-O-alkylguanine DNA alkyltransferase. Bioconjug Chem. 2009;20:1010–1015. doi: 10.1021/bc9000257. [DOI] [PubMed] [Google Scholar]
- 21.Niesen J, Stein C, Brehm H, et al. Novel EGFR-specific immunotoxins based on panitumumab and cetuximab show in vitro and ex vivo activity against different tumor entities. J Cancer Res Clin Oncol. 2015 doi: 10.1007/s00432-015-1975-5. [DOI] [PubMed] [Google Scholar]
- 22.Nachreiner T, Kampmeier F, Thepen T, et al. Depletion of autoreactive B-lymphocytes by a recombinant myelin oligodendrocyte glycoprotein-based immunotoxin. J Neuroimmunol. 2008;195:28–35. doi: 10.1016/j.jneuroim.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 23.Ribbert T, Thepen T, Tur MK, et al. Recombinant, ETA′-based CD64 immunotoxins: improved efficacy by increased valency, both in vitro and in vivo in a chronic cutaneous inflammation model in human CD64 transgenic mice. Br J Dermatol. 2010;163:279–286. doi: 10.1111/j.1365-2133.2010.09824.x. [DOI] [PubMed] [Google Scholar]
- 24.Stöcker M, Pardo A, Hetzel C, et al. Eukaryotic expression and secretion of EGFP-labeled annexin A5. Protein Expr Purif. 2008;58:325–331. doi: 10.1016/j.pep.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 25.Verhoven B, Schlegel RA, Williamson P. Mechanisms of phosphatidylserine exposure, a phagocyte recognition signal, on apoptotic T lymphocytes. J Exp Med. 1995;182:1597–1601. doi: 10.1084/jem.182.5.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krauss J, Arndt MA, Vu BK, et al. Targeting malignant B-cell lymphoma with a humanized anti-CD22 scFv-angiogenin immunoenzyme. Br J Haematol. 2005;128:602–609. doi: 10.1111/j.1365-2141.2005.05356.x. [DOI] [PubMed] [Google Scholar]
- 27.Arndt MA, Krauss J, Vu BK, et al. A dimeric angiogenin immunofusion protein mediates selective toxicity toward CD22+ tumor cells. J Immunother. 2005;28:245–251. doi: 10.1097/01.cji.0000161396.96582.10. [DOI] [PubMed] [Google Scholar]
- 28.Hendrzak JA, Wallace PK, Morahan PS. Optimizing the detection of cell surface antigens on elicited or activated mouse peritoneal macrophages. Cytometry. 1994;17:349–356. doi: 10.1002/cyto.990170412. [DOI] [PubMed] [Google Scholar]
- 29.Leonidas DD, Shapiro R, Allen SC, et al. Refined crystal structures of native human angiogenin and two active site variants: implications for the unique functional properties of an enzyme involved in neovascularisation during tumour growth. J Mol Biol. 1999;285:1209–1233. doi: 10.1006/jmbi.1998.2378. [DOI] [PubMed] [Google Scholar]
- 30.Dickson KA, Kang DK, Kwon YS, et al. Ribonuclease inhibitor regulates neovascularization by human angiogenin. Biochemistry. 2009;48:3804–3806. doi: 10.1021/bi9005094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stahnke B, Thepen T, Stocker M, et al. Granzyme B-H22(scFv), a human immunotoxin targeting CD64 in acute myeloid leukemia of monocytic subtypes. Mol Cancer Ther. 2008;7:2924–2932. doi: 10.1158/1535-7163.MCT-08-0554. [DOI] [PubMed] [Google Scholar]
- 32.Schiffer S, Letzian S, Jost E, et al. Granzyme M as a novel effector molecule for human cytolytic fusion proteins: CD64-specific cytotoxicity of Gm-H22(scFv) against leukemic cells. Cancer Lett. 2013;341:178–185. doi: 10.1016/j.canlet.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 33.Dunphy CH, Tang W. The value of CD64 expression in distinguishing acute myeloid leukemia with monocytic differentiation from other subtypes of acute myeloid leukemia: a flow cytometric analysis of 64 cases. Arch Pathol Lab Med. 2007;131:748–754. doi: 10.5858/2007-131-748-TVOCEI. [DOI] [PubMed] [Google Scholar]
- 34.Santos IM, Franzon CM, Koga AH. Laboratory diagnosis of chronic myelomonocytic leukemia and progression to acute leukemia in association with chronic lymphocytic leukemia: morphological features and immunophenotypic profile. Rev Bras Hematol Hemoter. 2012;34:242–244. doi: 10.5581/1516-8484.20120058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schiffer S, Rosinke R, Jost E, et al. Targeted ex vivo reduction of CD64-positive monocytes in chronic myelomonocytic leukemia and acute myelomonocytic leukemia using human granzyme B-based cytolytic fusion proteins. Int J Cancer. 2014;135:1497–1508. doi: 10.1002/ijc.28786. [DOI] [PubMed] [Google Scholar]
- 36.Karehed K, Dimberg A, Dahl S, et al. IFN-gamma-induced upregulation of Fcgamma-receptor-I during activation of monocytic cells requires the PKR and NFkappaB pathways. Mol Immunol. 2007;44:615–624. doi: 10.1016/j.molimm.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 37.Wei H, Bera TK, Wayne AS, et al. A modified form of diphthamide causes immunotoxin resistance in a lymphoma cell line with a deletion of the WDR85 gene. J Biol Chem. 2013;288:12305–12312. doi: 10.1074/jbc.M113.461343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Conway O’Brien E, Prideaux S, Chevassut T. The epigenetic landscape of acute myeloid leukemia. Adv Hematol. 2014;2014:103175. doi: 10.1155/2014/103175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jankowska AM, Makishima H, Tiu RV, et al. Mutational spectrum analysis of chronic myelomonocytic leukemia includes genes associated with epigenetic regulation: UTX, EZH2, and DNMT3A. Blood. 2011;118:3932–3941. doi: 10.1182/blood-2010-10-311019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schnerch D, Yalcintepe J, Schmidts A, et al. Cell cycle control in acute myeloid leukemia. Am J Cancer Res. 2012;2:508–528. [PMC free article] [PubMed] [Google Scholar]
- 41.Scholl C, Gilliland DG, Frohling S. Deregulation of signaling pathways in acute myeloid leukemia. Semin Oncol. 2008;35:336–345. doi: 10.1053/j.seminoncol.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 42.Gao X, Xu Z. Mechanisms of action of angiogenin. Acta Biochim Biophys Sin (Shanghai) 2008;40:619–624. doi: 10.1111/j.1745-7270.2008.00442.x. [DOI] [PubMed] [Google Scholar]
- 43.Lu Z, Xu S. ERK1/2 MAP kinases in cell survival and apoptosis. IUBMB Life. 2006;58:621–631. doi: 10.1080/15216540600957438. [DOI] [PubMed] [Google Scholar]
- 44.Peng Y, Li L, Huang M, et al. Angiogenin interacts with ribonuclease inhibitor regulating PI3K/AKT/mTOR signaling pathway in bladder cancer cells. Cell Signal. 2014;26:2782–2792. doi: 10.1016/j.cellsig.2014.08.021. [DOI] [PubMed] [Google Scholar]
- 45.Sadagopan S, Veettil MV, Chakraborty S, et al. Angiogenin functionally interacts with p53 and regulates p53-mediated apoptosis and cell survival. Oncogene. 2012;31:4835–4847. doi: 10.1038/onc.2011.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ivanov P, Emara MM, Villen J, et al. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol Cell. 2011;43:613–623. doi: 10.1016/j.molcel.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.