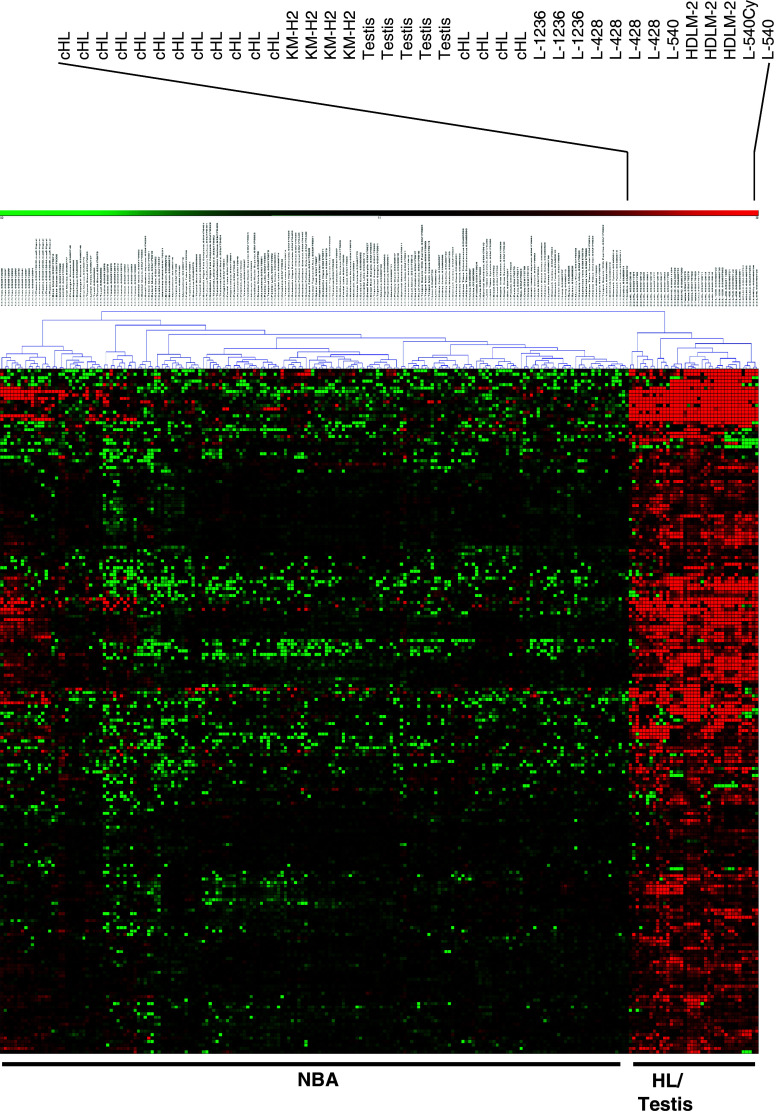
Fig. 4.
Expression of putative CTA in HL cells (HG_U133Plus2.0 analysis). Putative HL-associated CTA were identified by using MAFilter software as described in the text. The following GEO data sets have been used: GSE12453, GSE7307, GSE3526, GSE14879, GSE20011, GSE25986 and GSE12427. Presented is a cluster analysis with the identified probe sets (log2-transformed and median-centered signal intensities, absolute Manhattan distance and complete linkage clustering). Cluster analysis was performed with Genesis. The positions from samples from HL cell lines, micro-dissected HL cells (cHL), testes and other normal samples (NBA) are indicated