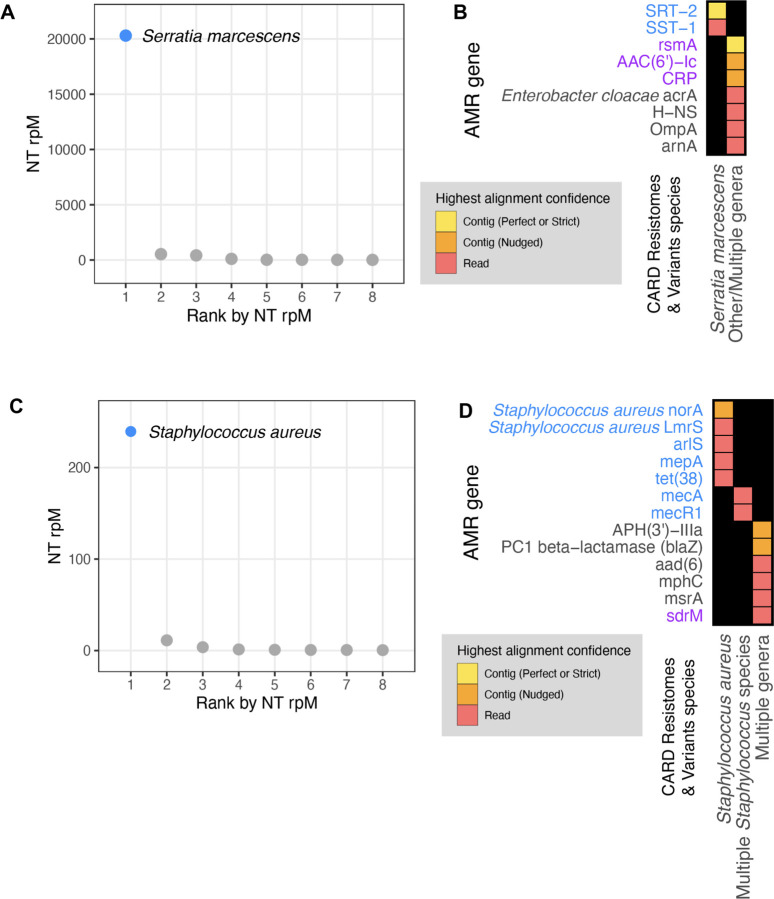
Figure 6: Co-detection of microbes and AMR genes in patients with critical bacterial infections using the CZ ID mNGS and AMR modules.
(A) Relative abundance (reads per million, rpM) of the eight most abundant taxa in the lower respiratory tract detected by RNA mNGS of tracheal aspirate from a patient with Serratia marcescens pneumonia. The dominant microbe is highlighted in blue. (B) AMR genes and their species prediction by the AMR module. Columns indicate the species these AMR genes and their variants are found in according to CARD Resistomes & Variants database, and those found in the dominant species as in (A) are colored in blue. AMR genes that are further associated with the dominant species by the pathogen-of-origin analysis are colored in purple. (C) Relative abundance (rpM) of the eight most abundant taxa detected by plasma DNA mNGS in a patient with sepsis due to MRSA bloodstream infection. The dominant microbe is highlighted in blue. (D) AMR genes and their species prediction by the AMR module. Columns indicate the species these AMR genes and their variants are found in according to CARD Resistomes & Variants database, and those found in the dominant species as in (C) are colored in blue. AMR genes that are further associated with the dominant species by the pathogen-of-origin analysis are colored in purple.