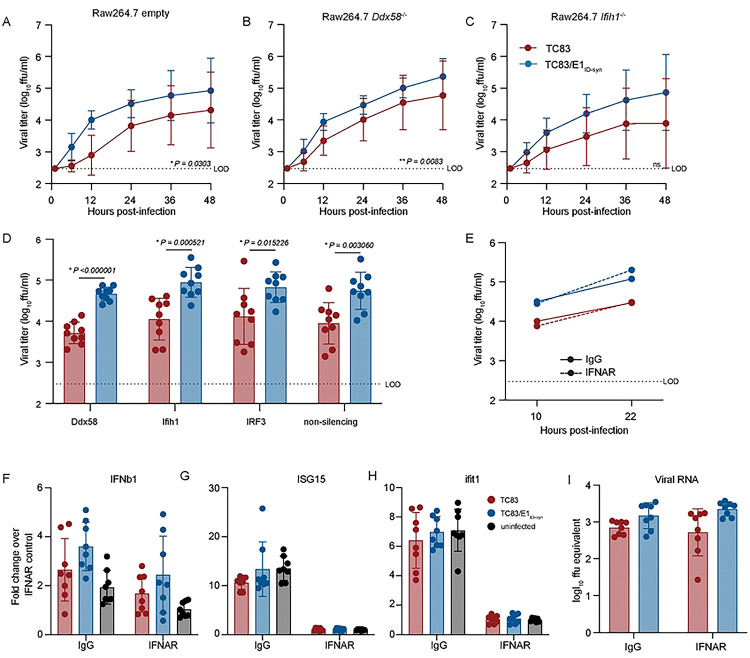
Figure 3. Differential macrophage replication of TC83 and TC83/E1ID-syn viruses is IFN- and RLR-independent.
(A-C) Replication kinetics of VEEV TC83 and TC83/E1ID-syn in (A) empty vector, (B) Ddx58−/−, and (C) Ifih1−/− CRISPR Raw264.7 cells. Cells were infected with indicated viruses at a MOI of 0.1. Cell culture supernatant was serially harvested at 1, 6, 12, 24, 36, and 48 hpi and infectious virus was titered using focus forming assay (FFA). Each experiment was performed in triplicate three times independently and the mean and SD are graphed. Statistical analysis was performed by calculating the area under the curve (AUC) for each replicate and experiment, and the AUC values for each virus analyzed by unpaired t-test. (D) Raw264.7 were treated with non-silecing control (NSC) siRNA or siRNA targeting Mavs, Ddx58, Ifih1, or Irf3. Cell culture supernatants were harvested at 24 hpi and infectious virus quantified by FFA. (E) Raw264.7 were pretreated for 1 hour with 10μg of IgG or IFNAR blocking antibody, then infected with TC83 or TC83/E1ID-syn at an MOI of 0.1 in the presence of antibody. Infectious virus from cell culture supernatants harvested at 10 and 22 hpi was titered by FFA. Each experiment was performed three times independently. (F, G) IFNAR blocking antibody assays were performed in WT Raw264.7 as described in E, and cell lysates collected at 22 hpi. IFNb1, ISG15, Ifit1 and VEEV viral RNA transcripts quantified by qRT-PCR. Gene expression within samples was normalized to hprt, and fold change in gene expression relative to IFNAR samples was calculated. Each experiment was performed three times independently in duplicate or triplicate, and statistical analysis was performed using unpaired t-test.