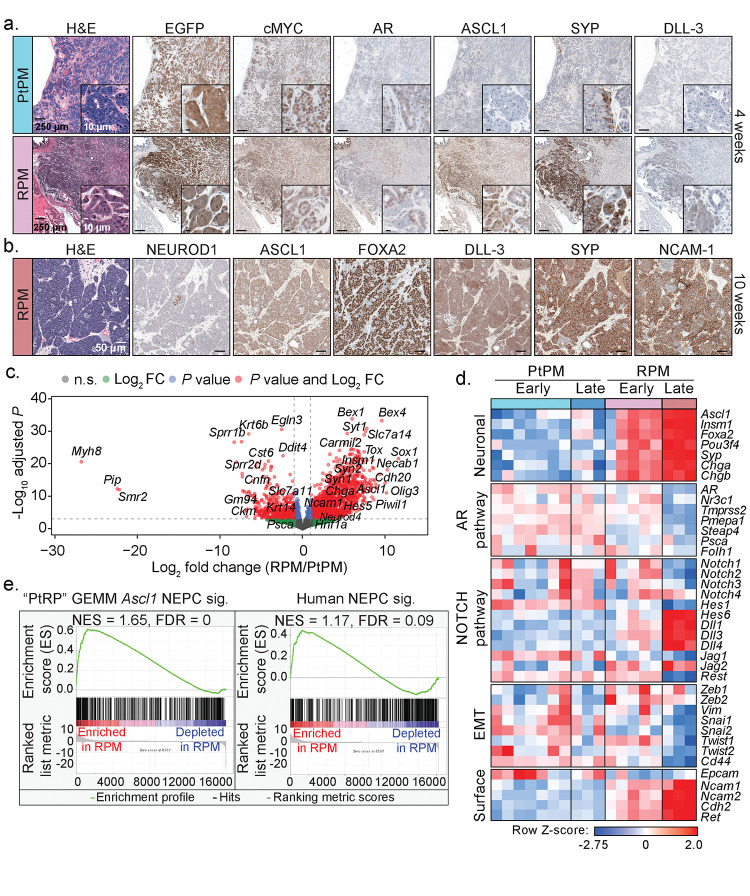
Figure 2: Molecular characterization of PtPM and RPM primary prostate tumor transplants demonstrates emergence of neuroendocrine carcinoma marker expression.
a. Representative histological analysis of PtPM (top) and RPM (bottom) tumors isolated at 4-weeks post-transplantation. Serial sections depict immunohistochemical staining of the indicated markers. Data are representative of n=22 independently transplanted tumors. b. Representative histological analysis of RPM tumors isolated at 10-weeks post-transplantation. Serial sections depict immunohistochemical staining of the indicated markers. Data are representative of n=25 independently transplanted tumors. c. Volcano plot depiction of the log2 fold change in RNA expression of primary (OT) RPM tumors relative to primary (OT) PtPM tumors. Genes that meet or surpass the indicated thresholds of significance and fold change in expression are color coded as depicted in the figure legend. Data derived from the comparison of PtPM (n=10) and RPM (n=8) independent prostate tumors. d. Heatmap depicting the Z-score normalized differential expression of select genes in PtPM versus RPM tumors. Time points of isolation are color coded in the figure as they are in Fig. 2a. Genes are grouped by the listed class or pathway. Early PtPM ≤4 weeks, early RPM ≤6 weeks. Late PtPM =5 weeks, late RPM =10 weeks. Data related to samples used in Fig. 2f. e. Enrichment plots (GSEA) of established expression signatures of (left) genetically engineered mouse model (GEMM) of NEPC harboring conditional deletion of Pten, Rb1, and Trp53 (PtRP), and (right) histologically verified human NEPC within RPM primary tumors. FDR and NES indicated in the figure. Analysis derived from the transcriptional profiles of multiple independent RPM tumors (n=8) relative to PtPM tumors (n=10). Data related to samples used in Fig. 2d. All scale bars noted in each panel and are of equivalent magnification across each marker.