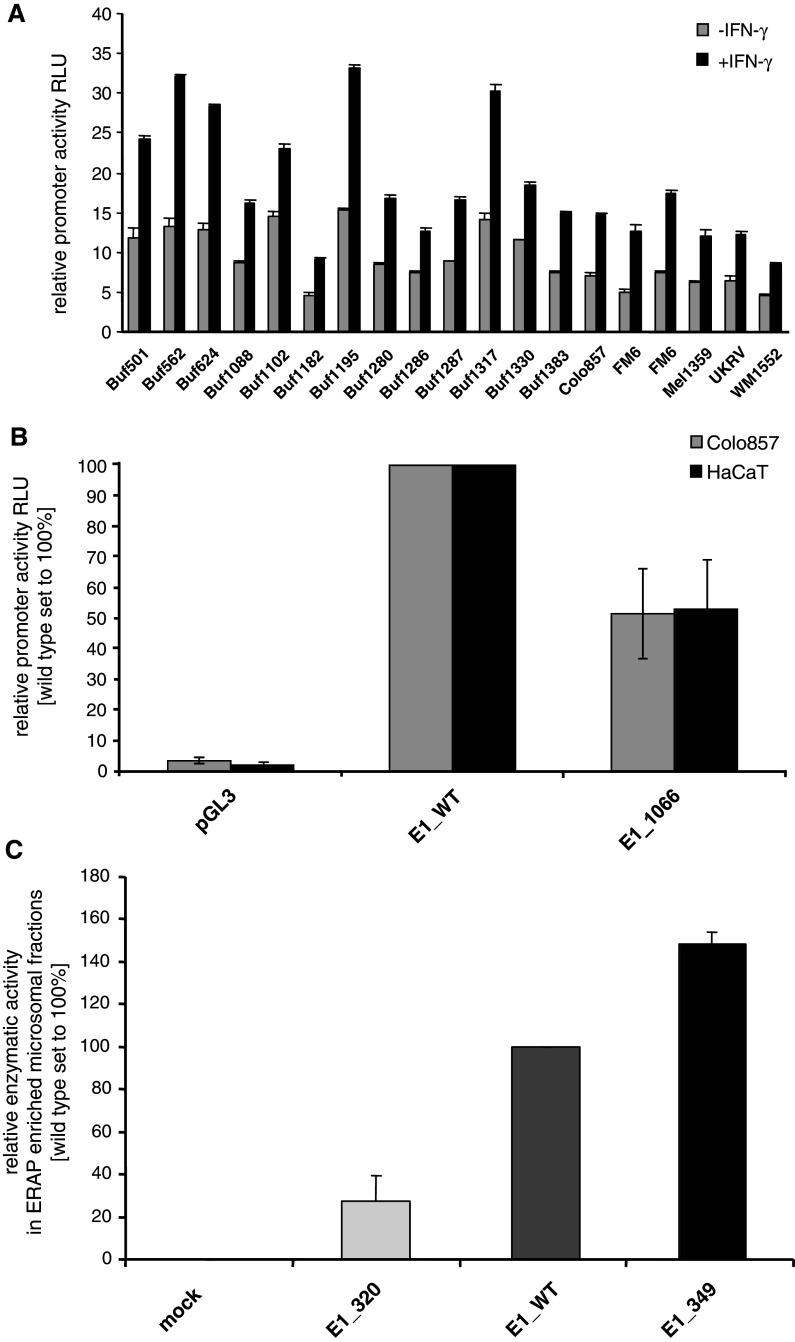
Fig. 2.
Heterogeneous ERAP promoter activity in melanoma cells. a The wt ERAP1 promoter and the pGL3 enhancer vector, which served as a control, were transiently transfected into a series of melanoma cells as indicated on the x-axis. Cells were left untreated or treated for 24 h with IFN-γ before the given ERAP1 (a) promoter activities were determined as described in “Materials and methods”. All results are expressed as relative luciferase activity (RLU) normalized to the corresponding β-galactosidase activity. Grey bars represent untreated cells; black bars IFN-γ-treated cells. b The relative activities of the wt ERAP1 (E1_WT) and mutated ERAP1 promoter (E1_1066) constructs as well as of the corresponding mock control (pGL3 enhancer vector) were transfected in the melanoma cell line Colo857 and the keratinocyte cell line HaCaT serving as a control. The relative promoter activities are expressed as RLU as outlined above in a. The mutation located at nt position 1066 leads to a markedly reduced transcriptional activity, independent from the cellular background. c Altered aminopeptidase activities of ERAP1 variants. The relative enzymatic activities of various ERAP1 constructs in the microsome fractions were determined as described in “Materials and methods”. The ERAP1-negative cell line Buf1182 was either transfected with the empty vector (mock), the wtERAP1 (E1_WT), the novel mut349ERAP1 (E1_349) or with the previously described enzymatically almost inactive ERAP1 variant mut320ERAP1 (E1_320). The relative enzymatic activities are expressed as RLU normalized to the relative ERAP1 enrichment within the indicated microsomal fractions as determined by ERAP1-targeting Western blot analyses. The newly characterized variant with the amino acid substitution M→V at position 349, which is located near the active site, slightly enhances the enzymatic activity of ERAP1