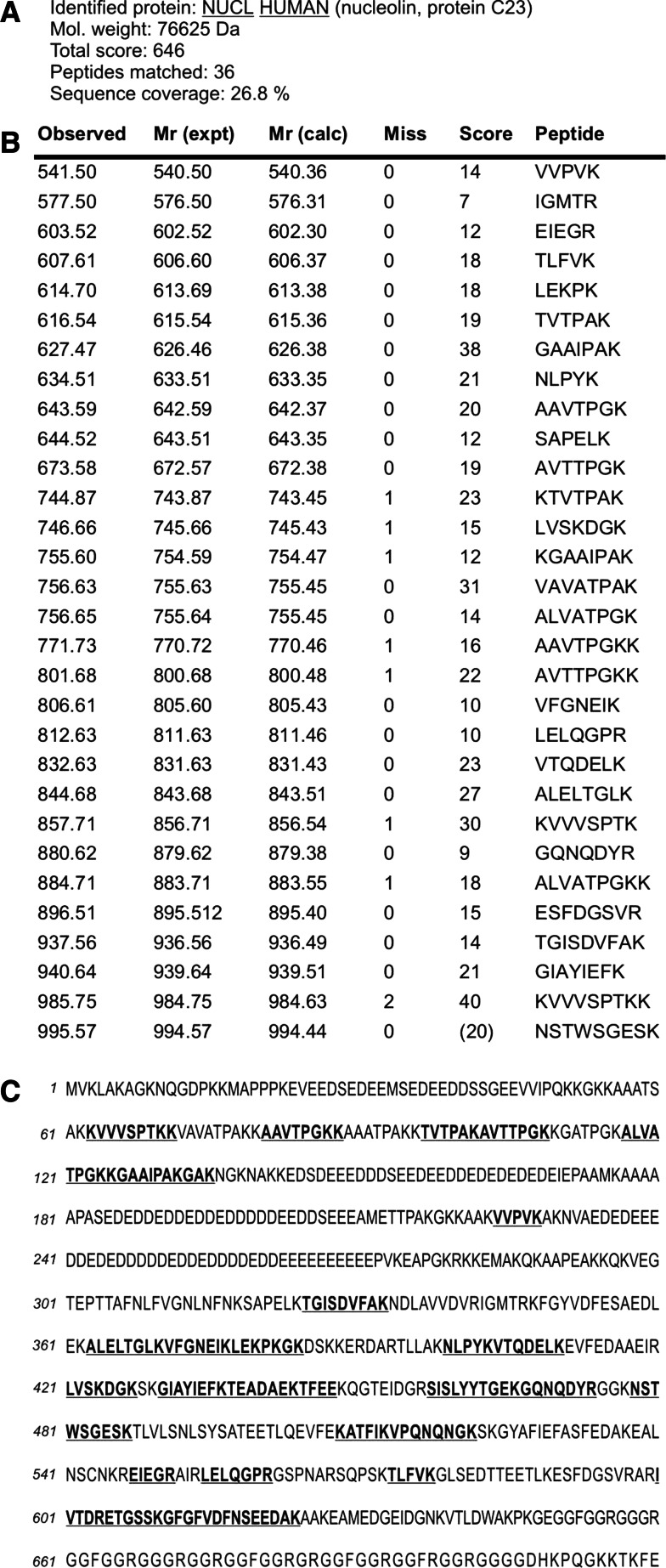
Fig. 3.
Results of the mass spectrometry analysis of in-gel tryptic digest of 105 kDa protein band isolated from total lysate of Ma-Mel-27 cells using VVL-agarose. The encircled band shown in Fig. 2 lane 7 was excised from polyacrylamide gel and digested with trypsin, and the tryptic peptides were sequenced. The acquired spectra were analysed using Bruker Data Analysis software and interpreted using Mascot search engine against Swiss-Prot/TrEMBL sequence database. Peptides determined by sequencing were found to correspond to a high degree of certainty to human nucleolin (a). The table columns contain: Observed, experimental m/z value; Mr(expt), experimental m/z transformed to a relative molecular mass; Mr(calc), relative molecular mass calculated from the matched peptide sequence; Miss, number of missed cleavage sites; Score, ions score; Peptide, sequence of the matched peptide in 1-letter code (b). Localization of the identified peptides within the nucleolin sequence. The primary structure of human nucleolin is shown in the single-letter amino acid code sequence. Matched tryptic peptides are in bold and underlined (c). Note. Matched peptides equally cover the nucleolin sequence