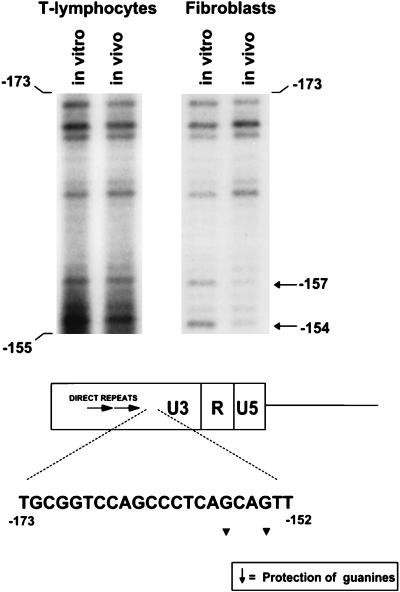
FIG. 5.
In vivo DMS footprinting in the GC-rich sequences. The regions from DMS footprinting gels analogous to those shown in Fig. 2 and 4 and corresponding to the GC-rich sequences downstream of the 75-bp repeats are shown for thymic tumor DNA and 43-D infected fibroblasts. Evidence for protein interactions at positions −153 and −155 in the sequence CAGCAG was obtained for fibroblasts but not for M-MuLV-infected Ti-6 cells or the primary thymic tumor cells. Infected Ti-6 cells showed the same pattern as the thymic tumor cells (no protection [data not shown]). The relative positions of the protected bases in the GC-rich sequences are shown.