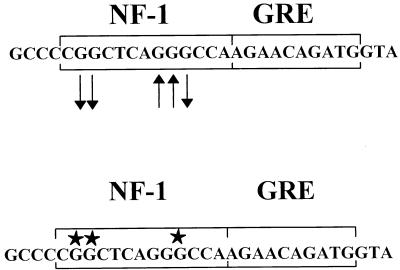
FIG. 7.
Comparison of in vivo footprints and in vitro methylation interference results. Nuclear protein interactions at the NF-1 site reported by Speck and Baltimore (40) detected by methylation interference with WEHI 231 nuclear extracts are shown in the lower sequence (stars indicate G residues that interacted with protein). The results of in vivo DMS footprinting are shown in the upper sequence. Guanine bases protected in vivo (arrows pointing away from bases) correspond to guanines shown to be protein contact points by methylation interference.