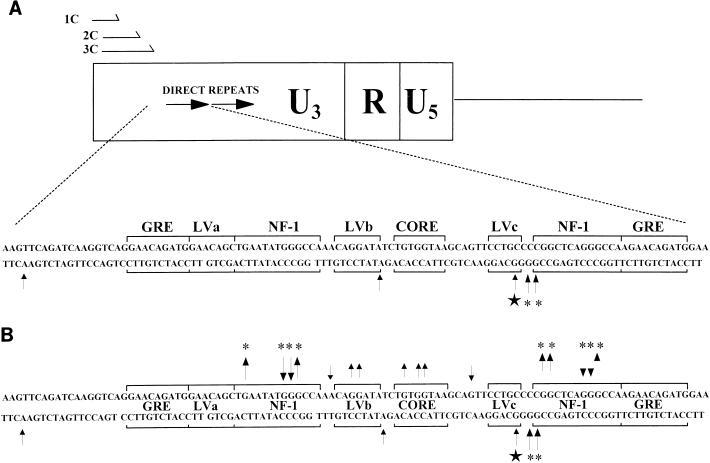
FIG. 8.
DNA-protein interactions on the minus strand obtained by in vivo footprinting. (A) The nested oligonucleotide primer set used for the minus-strand in vivo footprinting (1C, 2C, and 3C) is shown. A summary of the minus (lower)-strand bases hypersensitive to in vivo DMS methylation in fibroblasts and primary thymic tumor cells was compiled analogously to that shown in Fig. 6. These results were taken from the in vivo footprinting analysis shown in Fig. 9. Asterisks indicate bases hypersensitive to DMS methylation in infected fibroblasts, the star indicates a hypersensitive base specific to primary thymic tumor cells, and arrows indicate sites hyperactive to DMS. (B) A summary of in vivo protein-DNA interactions observed for both DNA strands is shown. Arrows pointing away from bases on either strand indicate DMS protection, while arrows pointing towards bases indicate DMS-hypersensitive sites. Note that protein interactions on the minus strand were evident only as hypersensitive sites. The data for the sense (upper) strand are specific for the upstream LTR, while the data for the minus strand are composites of the upstream and downstream LTRs. Stars indicate T-lymphoid-cell-specific bases, and asterisks indicate fibroblast-specific bases.