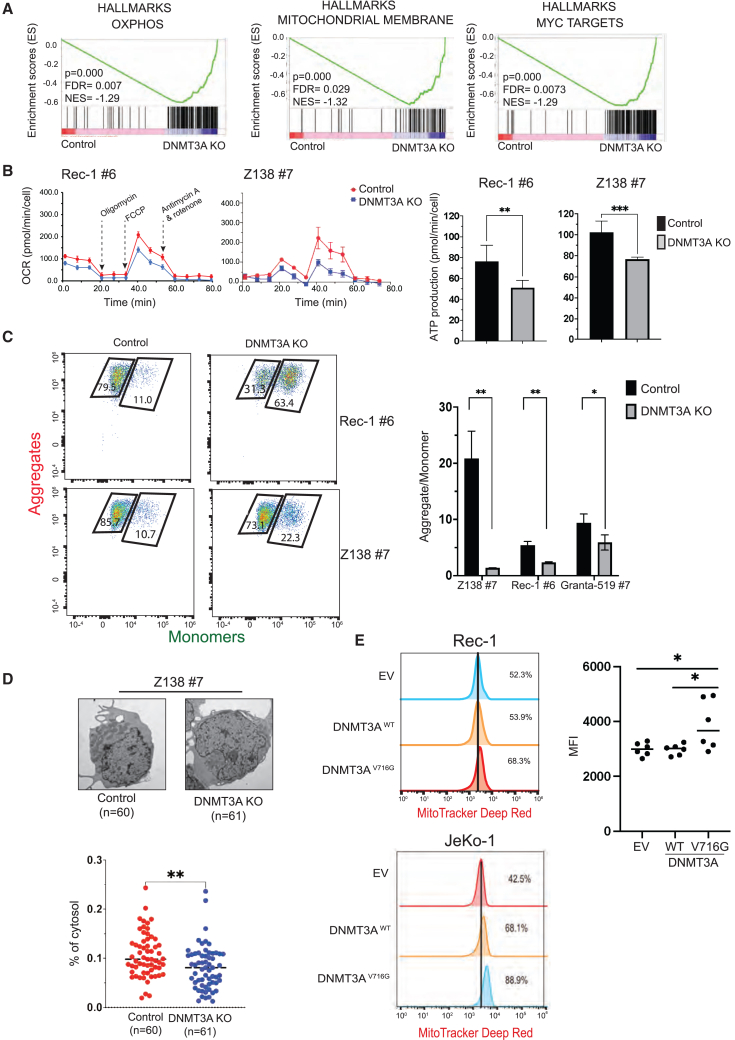
Figure 2.
DNMT3A expression is required for OXPHOS in MCL cells by controlling mitochondrial biogenesis independent of its methyltransferase activity
(A) GSEA of DNMT3A KO cells shows downregulation of genes involved in OXPHOS, mitochondrial biogenesis, and MYC transcriptional pathway compared to control cells.
(B) The reduced rate of OCR (pmol/min/cell) and ATP productions in DNMT3A KO cells relative to control cells. All of the OCR values were normalized to total live-cell number. OCR experiments were performed independently at least 2 times, each with 6 technical replicates.
(C) Reduced MMP in DNMT3A KO cells compared to control cells by flow cytometric analysis of JC-1 dye.
(D) Representative images of TEM of Z-138 no. 7 control cells compared to DNMT3A KO cells (top). A significant decrease in mitochondrial mass, approximated by the relative area of mitochondria to cytosol per cell, of DNMT3A KO cells relative to control cells (bottom). Each data point represents an individual cell (n = 60–61 cells). TEM experiments were performed independently twice, each with duplicates.
(E) DNMT3AV716G expression was sufficient to increase MMP in Rec-1 and JeKo-1 cells and the quantification of MMP in Rec-1 cells. Error bars represent mean ± SD (Student’s t test, 2 independent experiments in triplicate).