Figure 3.
Clonal dynamics following drug treatment
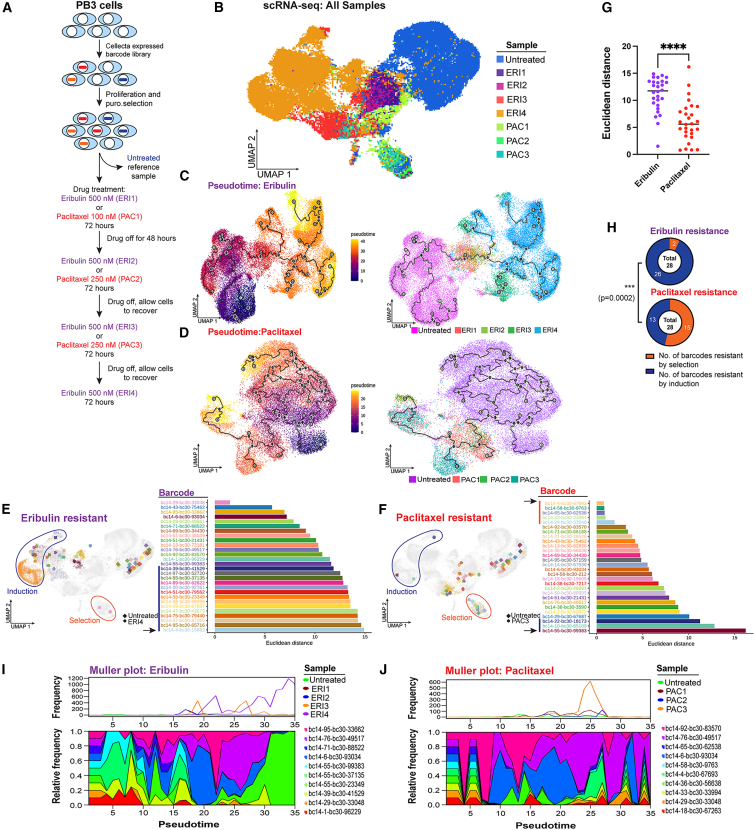
(A) Schematic outlining genetic barcoding of cells and subsequent drug treatment strategy.
(B) UMAP projection of scRNA-seq data obtained from PB3 cells before and after 1–4 rounds of treatment with eribulin or paclitaxel.
(C and D) Monocle pseudotime projection showing the trajectory of cancer cell evolution upon drug treatment. The main lineage path or master path/node is indicated by “1” in a white circle. Sublineages (leaf nodes) from the master path are indicated by numbers in gray circles. Branches from leaf nodes are indicated by numbers in black circles. Left and right: cells colored according to pseudotime and sample, respectively.
(E–G) Euclidean distance between ERI4 vs. untreated cells (E) or PAC3 vs. untreated cells (F), with the same barcode are shown in swimmer plots, and UMAPs represent median points of ERI4 or PAC3 and untreated cells. ∗∗∗∗p < 0.0001 by Mann-Whitney U test.
(H) Proportions of induction vs. selection resistance for ERI and PAC treatments were compared by chi-square test.
(I and J) Muller plots outlining clonal diversity of the 10 most abundant barcodes following drug treatment.