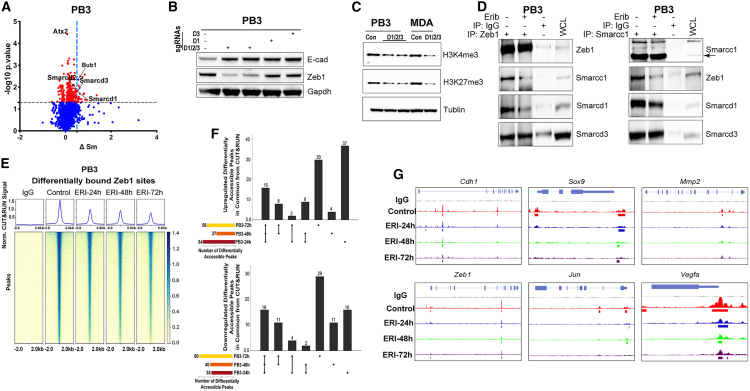
Figure 5.
ZEB1-SWI/SNF interactions are necessary for maintenance of a mesenchymal state
(A) PB3 cells treated with or without 300 nM eribulin for 4 h were analyzed by PISA assay. Proteins were plotted by ΔSm and −log10p (t test).
(B and C) Immunoblot analysis of cells with CRISPR-Cas9-mediated deletion of SmarcD1 (D1), SmarcD3 (D3), or SmarcD1/2/3.
(D) Zeb1 and Smarcc1 immunoprecipitates and lysates from cells treated with or without eribulin for 48 h.
(E–G) Genome-wide occupancy of Zeb1 in PB3 cells treated with or without eribulin for 24, 48, or 72 h was evaluated using CUT&RUN. In (E), average CUT&RUN enrichment profiles (top) and heatmap of the CUT&RUN signal ±2 kb of peaks (bottom) are shown. Shown in (F) are UpSet plots of differentially accessible peaks at the indicated time points as mean ± SEM. Signal tracks of Zeb1 localization at target gene loci are shown in (G).