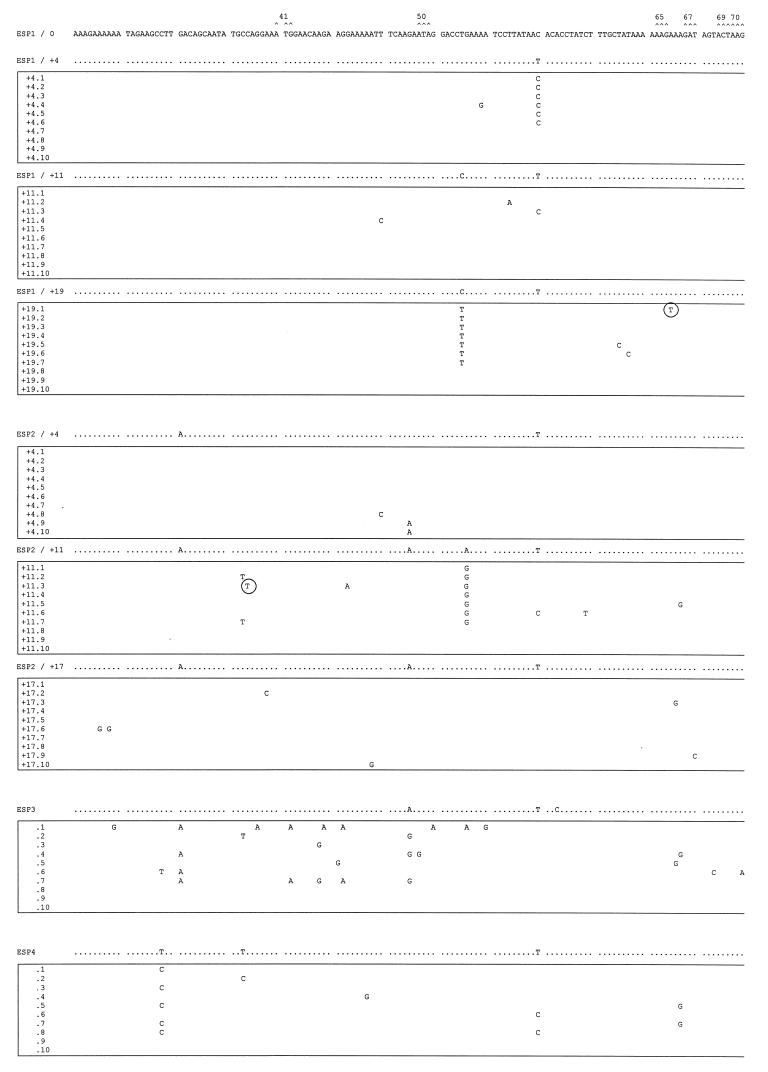
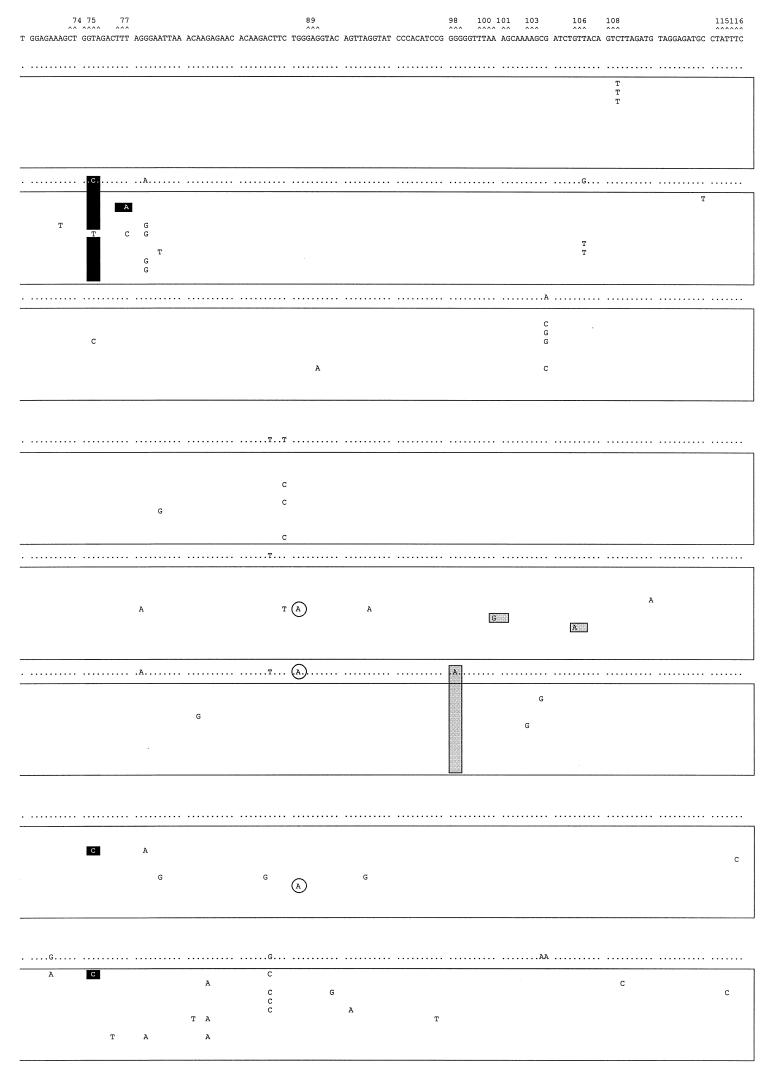
FIG. 5.
Mutant spectra of pol gene quasispecies in samples from the longitudinal study and the other two HIV-1 group O isolates. Nucleotide sequences (positions 82 to 348, encoding amino acids 28 to 116 of the RT) of clones from each sample are boxed. The uppermost sequence corresponds with the sample used as reference (ESP1/0). The nucleotide sequence at the top of each box represents the consensus nucleotides (determined experimentally by the direct sequencing of the average uncloned PCR product) for each sample. Only those nucleotides that differ from the ESP1/0 sequence are indicated. The mutant spectrum of each quasispecies is represented by the number of mutations from the corresponding consensus sequence. Triplets encoding residues involved in resistance to RT inhibitors (16) are numbered and marked (∧) above the ESP1/0 sequence. Shaded and solid boxes designate nucleotide mutations encoding amino acid substitutions associated with resistance to NNRTIs or nucleoside analogs, respectively. Encircled nucleotides are those mutations leading to stop codons in the pol gene.