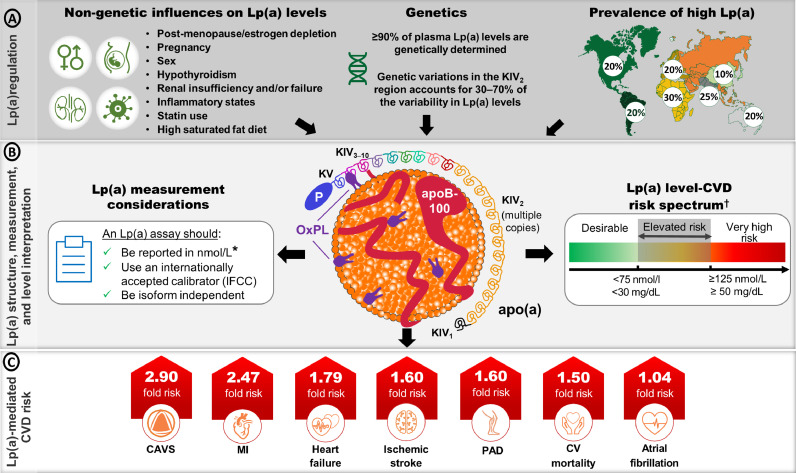
Fig. 1.
Overview of the structure, regulation, measurement considerations and level interpretation of Lp(a) and CVD-associated risk. A) Listing of non-genetic and genetic factors that modolute lipoprotein(a) [Lp(a)] levels. Lp(a) levels are primarily determined by variability in the LPA gene which codes for the apolipoprotein(a) [apo(a)] component of the particle. [5]. The prevalence of High Lp(a) levels ranges from 10-30% and varies depending on ancestry [13]. B) The Lp(a) particle comprises a single apolipoprotein B-100 (apoB-100) containing lipoprotein covalently associated with apo(a). Oxidized phospholipids (OxPLs) are covalently associated with apo(a), apoB-100 and the lipid core. Apo(a) consists of 10 kringle IV (KIV) domains, a single KV domain and an inactive, protease-like domain. Different apo(a) isoforms have different KIV2 copy numbers [69]. Recent guidance on important considerations when choosing Lp(a) assays, along with the risk thresholds for determining risk from an Lp(a) measurement are described [5,10]. C) Adjusted hazard ratios for select cardiovascular disease (CVD) outcomes comparing participants in the top percentile of Lp(a) distribution (>95th percentile for calcific aortic valve stenosis [CAVS], ischemic stroke and CV mortality; >99th percentile for myocardial infarction [MI] and heart failure[HF]) versus those with lower Lp(a) levels (<22nd percentile for CAVS; <34th percentile for MI and HF; <50th percentile for ischemic stroke and CV mortality) [45]. Peripheral artery disease (PAD) risk is based on an adjusted odds ratio for Lp(a) >66th percentile compared to <33rd percentile [45]. Atrial fibrillation risk is based on a Mendelian randomization analysis of the odds ratio per 50 nmol/L increase in Lp(a) [55].
*Lp(a) levels are still reported in mg/dL and therefore knowledge of whether the Lp(a) measurement was in nmol/L or mg/dL is crucial; †conversion between nmol/L and mg/dL is a gross estimate. IFCC = International Federation of Clinical Chemistry.