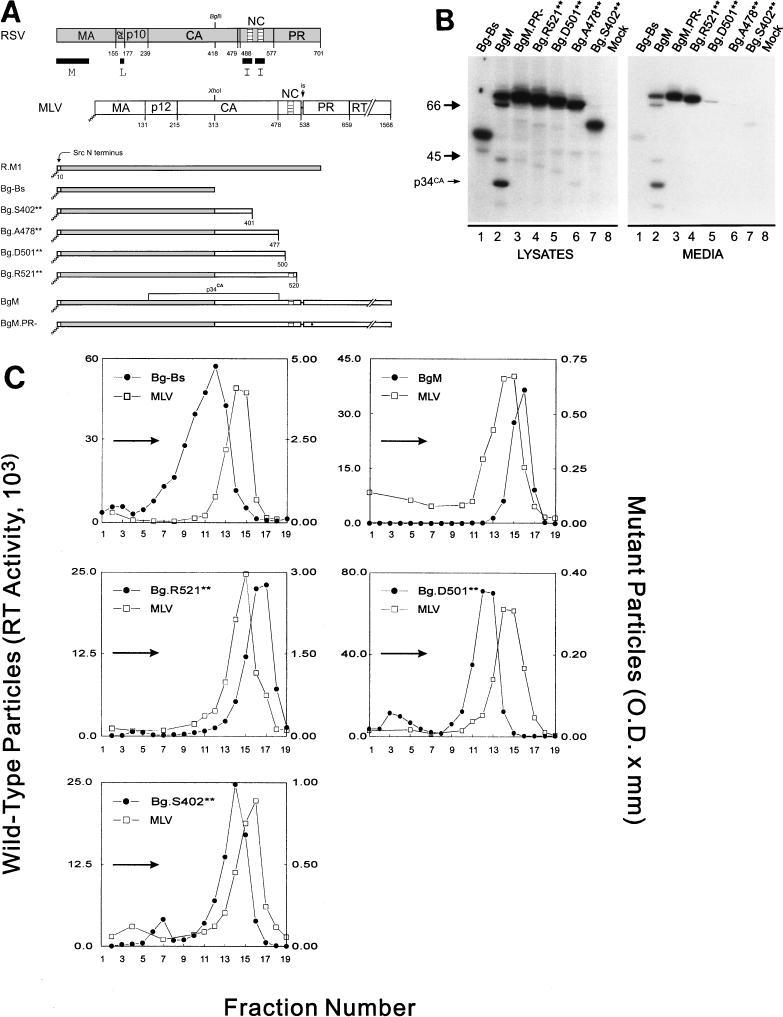
FIG. 1.
The C terminus of MLV contains at least one I domain. (A) Diagram of RSV-MLV chimeras. The wild-type RSV Gag (shaded) and MLV Gag (unshaded) proteins are aligned at the junction site of the chimeras (made by fusing the XhoI and BglII sites of the corresponding genes). The assembly domains of RSV are indicated by black boxes and the zinc fingers in NC by hatched regions. The site of in-frame suppression of the stop codon separating the MLV gag and pol genes is marked “is.” Sites of proteolytic cleavage are indicated by vertical lines through the proteins, and numbers refer to the last amino acid of the released products. The open box and squiggled line at the N terminus of the chimeras represent replacement of the first 10 RSV amino acids with those of pp60v-src and the consequent addition of myristic acid, respectively. The MLV protease active-site mutation is indicated by a black dot. Chimera R.M1 has been previously referred to as Myr1 (37). (B) Expression of RSV-MLV chimeras. The RSV-MLV constructs were transfected into COS-1 cells, and subsequently labeled for 2.5 h with l-[4,5-3H(N)]leucine. The Gag proteins from the medium and lysates of the transfected cells were immunoprecipitated with a mixture of α-RSV and α-MLV antibodies, separated by SDS-PAGE, and visualized by autoradiography. The numbers to the left are the positions (in kilodaltons) of the molecular mass markers, and the position of the RSV-MLV hybrid CA cleavage product p34CA is indicated. (C) Sucrose density gradient analysis. Cells were transfected and subsequently labeled for 8 h. Medium was collected, cleared of cellular debris, mixed with purified MLV, and centrifuged for 16 h at 83,500 × g through a 10 to 50% sucrose gradient. The gradients were fractionated, and the amount of control or experimental particles in each fraction was determined by reverse transcriptase assays or immunoprecipitation followed by densitometry of autoradiograms, respectively. An arrow in each panel indicates the direction of sedimentation. O.D., optical density.