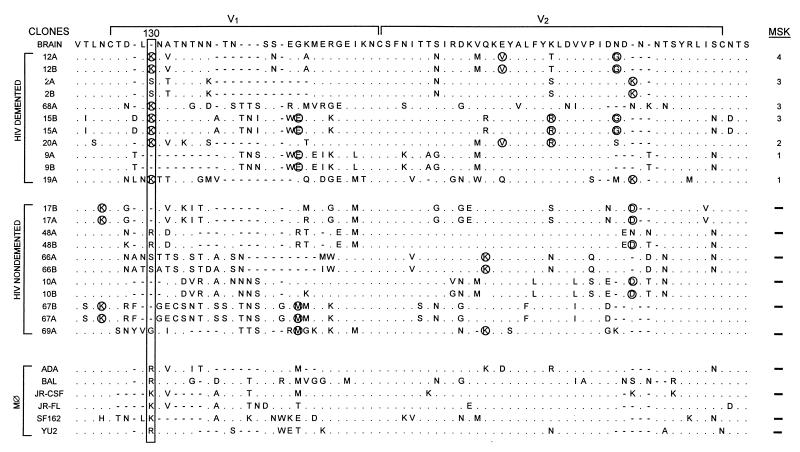
FIG. 3.
Brain-derived V1-V2 envelope sequences from HIV-D (D) and HIV-ND (ND) patients aligned with the brain consensus sequence and corresponding sequences from established macrophagetropic viruses. HIV-D sequences are presented in order of severity of dementia according to MSK score shown at the right. Two clones were sequenced from each patient except for patients 68, 20, 19, and 69. Position 130 (boxed) shows significant difference between HIV-D and HIV-ND groups (Fisher’s exact, P < 0.01), with a lysine predominating in the HIV-D group and a variable amino acid in the HIV-ND group. Comparison of the frequency of the Lys130 in established databases (38) revealed that it was detected significantly more frequently in the HIV-D sequences than in the database (Fisher’s exact, P < 0.01). Circled amino acids identified in two or more patients in one group, but not present in the other group at a specific position, were termed unique. The mean number of unique amino acids per clone ± SEM was significantly greater in the HIV-D group (3.0 ± 0.44) than in the HIV-ND group (1.6 ± 0.20) (Student’s t test, P < 0.05). In patients with two or more clones, the difference between clones varied from zero to three amino acids. The length of the V2 region, the number of positively charged amino acids, and number of potential glycosylation sites did not differ between HIV-D and HIV-ND groups.