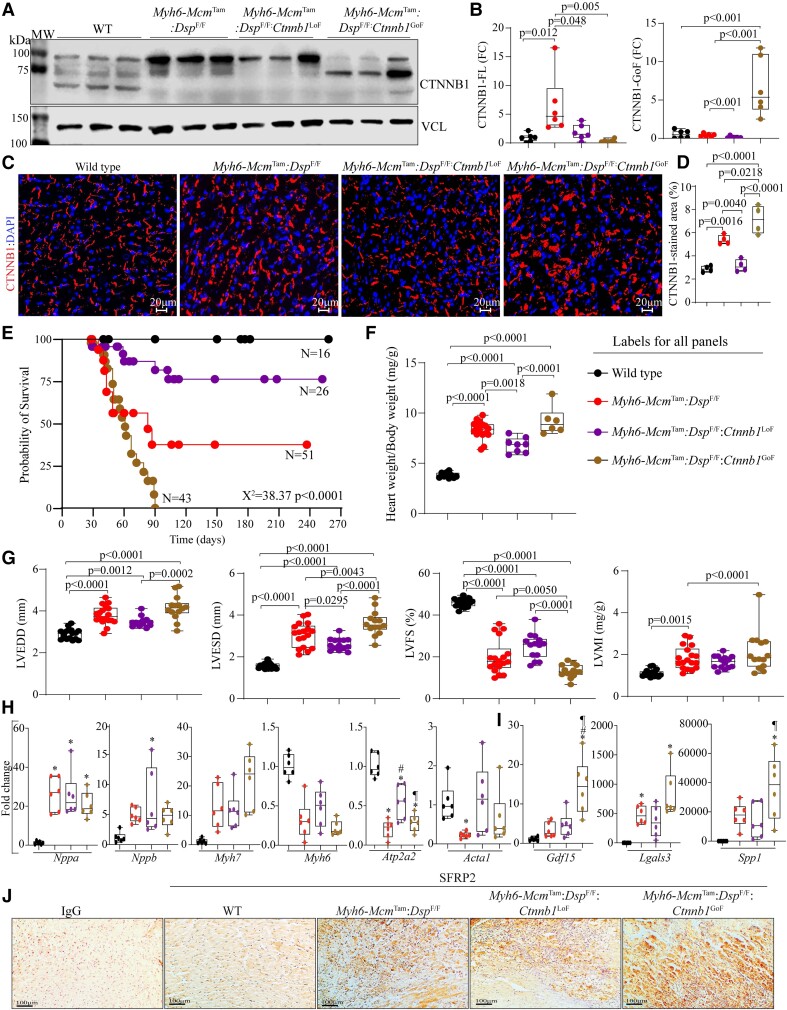
Figure 3.
Effects of genetic inactivation and activation of β-catenin on survival and cardiac function. (A) Immunoblot showing β-catenin expression in the WT, Myh6-McmTam:DspF/F, Myh6-McmTam:DspF/F:Ctnnb1LoF, and Myh6-McmTam:DspF/F:Ctnnb1GoF myocytes along with the vinculin (VCL), the latter as a control for loading conditions. (B) Quantitative data showing levels of full-length and stable β-catenin (exon 3 deleted, GoF) protein. (C and D) Immunofluorescence panels show expression and localization of the β-catenin in the myocardial sections along with the corresponding quantitative data. (E) Kaplan–Meier survival plots. (F) Heart weight/body weight ratio. (G) Selected echocardiographic indices, namely LV end-diastolic diameter (LVEDD), LV end-systolic diameter (LVESD), LV fractional shortening (LVFS), and LVMI. (H) Transcript levels of selected markers of hypertrophy and failure, as analysed by RT–PCR. (I) Transcript levels of selected secreted biomarkers of heart failure, as determined by RT–PCR. (J) Immunohistochemistry panels showing expression of SFRP2. Myocardial sections stained with Immunoglobulin G alone served as controls. The mean values among the groups were compared by ANOVA followed by Bonferroni correction for multiple comparisons. Only significant P values (Bonferroni corrected) are depicted in the panels throughout all figures. Sample size: Each dot in the panels represents one independent sample throughout the figures. Whenever a membrane is probed for multiple test proteins, the same blot for the loading control is included in the figures. The same color symbols in all quantitative panels are used for consistency and only one set of symbols is listed in each panel to avoid redundancy. * denotes P < 0.05 compared to WT, # P < 0.05 compared to Myh6-McmTam:DspF/F and ¶ P < 0.05 compared to Myh6-McmTam:DspF/F:Ctnnb1LoF.