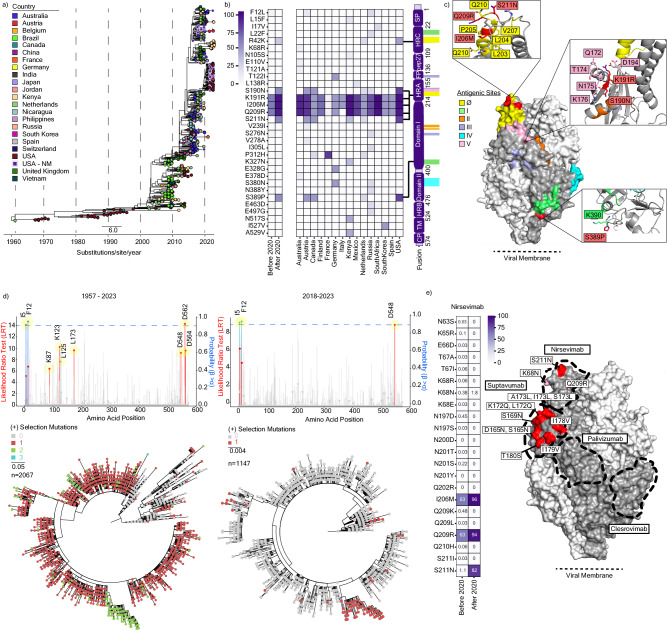
Fig. 5. Globally circulating RSV-B mutations in fusion protein.
a Maximum likelihood (ML) phylogenetic temporal analysis of globally circulating RSV-B whole genome sequences collected from 1957 to 2023 (as of October 10, 2023) (n = 723). Branch tips are colored by country of origin, and scale units are substitutions/site/year. b Heatmap of RSV-B Fusion mutational frequency among sequences sampled before and after May 1, 2020 relative to the GB5.0.5a most recent common ancestor (MRCA, left). Isolates sampled after May 1, 2020, are further stratified by country (n = 15, middle). A schematic of F domains with antigenic site positions highlighted is shown on the right. c Structure homology modeling of RSV-B prefusion F from the NMH/US cluster using template PDB:6QOS via SWISS-MODEL. Antigenic sites (yellow = Ø, green = I, orange = II, purple = III, cyan = IV, pink = V) and mutations defining the NMH/US cluster (red) are annotated on the trimer structure. Side chain residues within 5 angstroms of the three US cluster-defining mutations (S190N, S211N, S389P) are labeled in the insets. d Overlayed lollipop and bar plots for episodic and pervasive positive selection analysis, respectively, of unique RSV-B F open reading frames (ORFs) from 1957 to 2023 (n = 2067, right) and from 2018 to 2023 (n = 1147, left). Lollipop plots depict the likelihood ratio test (LRT) for episodic positive selection by a mixed effects model of evolution (MEME); significant positions are highlighted in red (p < 0.05). Overlaid bar charts depict the posterior probability of pervasive positive selection by Fast Unconstrained Bayesian AppRoximation (FUBAR); significant positions at greater than 90% are highlighted in blue. Corresponding ML phylogenetic trees (bottom) display the number of positions undergoing positive selection in a given branch. e Heatmap displaying mutational frequencies at positions within the Nirsevimab binding site before and after May 1, 2020 (left). Mutational frequency by position after May 1, 2020, is overlaid on the RSV-B prefusion F homology model (right). Proposed monoclonal antibody binding sites for Nirsevimab, Suptavumab, Clesrovimab, and Palivizumab are outlined.