FIGURE 1.
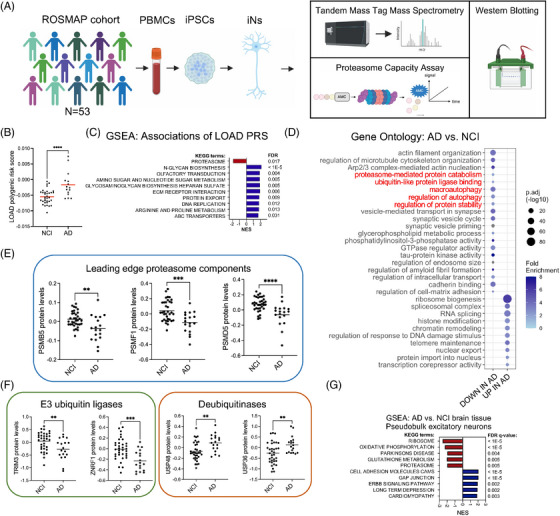
Genetically encoded proteasome vulnerability in Alzheimer's disease (AD) implicated in the analyses of induced pluripotent stem cell (iPSC) ‐derived neurons from over 50 individuals. (A) Overview of the study. (B) Polygenic risk scores (PRS) for late‐onset AD (LOAD) (n = 18) and not cognitively impaired (NCI) (n = 35) individuals in the ROSMAP (Religious Order Study or Memory and Aging Project) cohort examined in this study. Red bars represent the mean. Statistical analysis used unpaired t‐tests. (C) Gene set enrichment analysis (GSEA) using the KEGG (Kyoto Encyclopedia of Genes and Genomes) database of proteins correlated with LOAD PRS (by Pearson correlation analysis) in the ROSMAP iPSC‐derived neurons (iNs). (D) Gene ontology analysis of differentially expressed proteins in AD versus not cognitively impaired (NCI) individuals; adjusted p‐value < 0.05. Terms relating to protein turnover are highlighted in red. (E‐F) Protein expression of PSMB5, PSMF1, PSMD5, TRIM3, ZNRF1, USP48, and USP36 in NCI (n = 35) or AD (n = 18) individuals. Black bars represent the mean. Statistical analysis used unpaired t‐tests. (G) Single nucleus (sn)RNAseq data available on the AMP‐AD Knowledge Portal from human brain tissue (DLPFC [dorsolateral prefrontal cortex], BA9) from 131 AD and 162 NCI ROSMAP participants was used to compare pseudobulk RNAseq data between AD and NCI excitatory neurons. Shown are the results of GSEA of genes up and downregulated in AD versus NCI excitatory neurons, graphing only the top 5 terms in each direction. For comparisons in B, E, F: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
