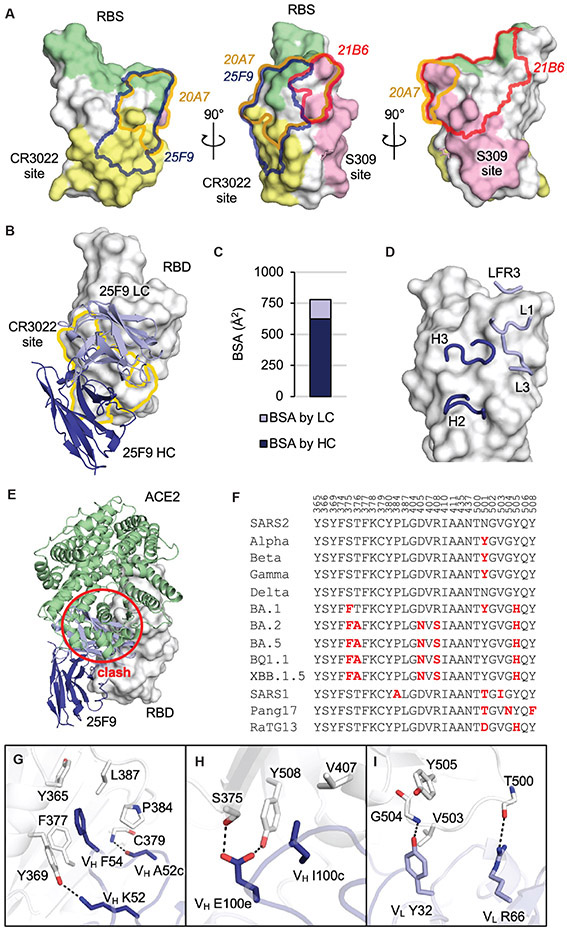
Fig. 3. 25F9 recognizes a conserved region on SARS-CoV-2 RBD.
The SARS-CoV-2 RBD is shown in white and human ACE2 is in pale green throughout all the figures; the heavy and light chains of 25F9 are in blue and lavender, respectively. For clarity, only variable domains of the antibodies are shown in all figures. (A) Shown are the relative positions of epitopes on SARS-CoV-2 RBD (white). The RBS is shown in pale green, the CR3022 site is shown in yellow, and the S309 site is shown in pink. Epitopes of 25F9, 20A7, and 21B6 are highlighted in blue, orange, and red outlines, respectively. RBS and epitope residues are defined as buried surface area (BSA > 0 Å2) as calculated by Proteins, Interfaces, Structures and Assemblies (PISA, http://www.ebi.ac.uk/pdbe/prot_int/pistart.html). (B) Shown is the crystal structure of 25F9 in complex with SARS-CoV-2 RBD. (C) The surface area of SARS-CoV-2 buried by heavy and light chains of 25F9 is shown. (D) 25F9 interacts with RBD using CDRs H2, H3, L1, and L3, as well as LFR3. (E) SARS-CoV-2 RBD with 25F9 superimposed onto an RBD-ACE2 complex structure (PDB 6M0J) shows that 25F9 would clash (indicated with a red circle) with ACE2. (F) Shown is sequence alignment of epitope residues in a subset of SARS-like viruses. Residues that differ from wild-type SARS-CoV-2 are indicated in red. SARS2, wild-type SARS-CoV-2; SARS1, SARS-CoV-1. (G to I) Molecular interactions between RBD and CDR H2 (G), CDR H3 (H), and light chain (I) are shown. Hydrogen bonds and salt bridges are indicated by dashed lines.