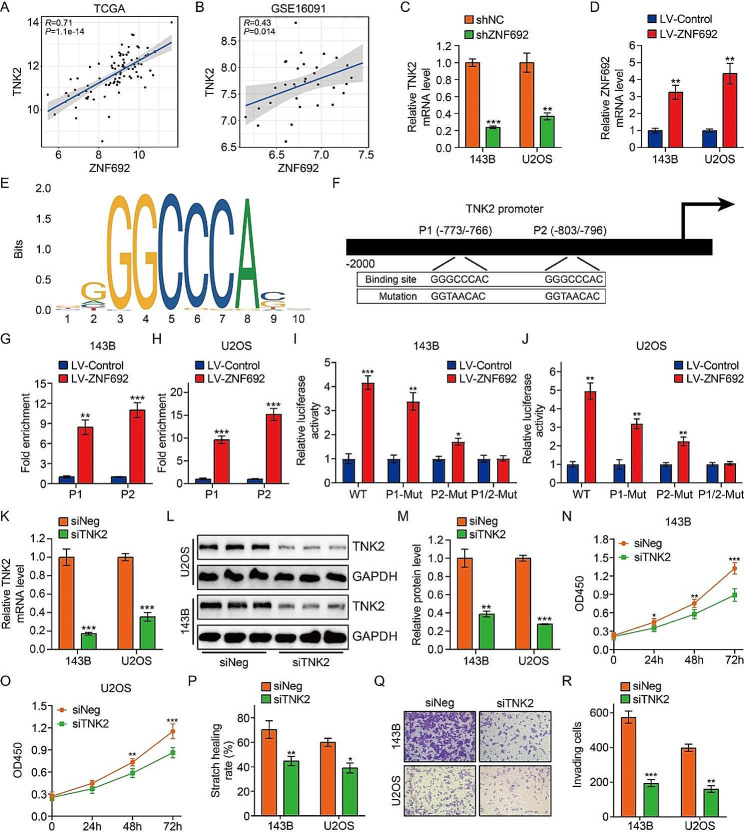
Fig. 5.
Identification of TNK2 as a direct transcriptional target of ZNF692. (A-B) ZNF692 expression was significantly positively correlated with the expression of TNK2 based on TCGA and GSE16091 datasets. (C-D) qRT-PCR analysis of TNK2 mRNA levels in 143B and U2OS cells stably knocking down or overexpressing ZNF692. (E) The binding motif of ZNF692 predicted by JASPAR online tool. (F) The two putative ZNF692-binding sites (P1 and P2) in the − 2000 bp to 0 bp sequence upstream of the transcription start site of TNK2. (G-H) ChIP-qPCR assays in 143B and U2OS cells stably overexpressing ZNF692 confirmed significant enrichment of ZNF692 in the two putative binding sites of TNK2. (I-J) A luciferase reporter assay suggested that ZNF692 apparently forced TNK2 promoter activity. Site-specific mutagenesis was used to determine ZNF692-responsive regions in the TNK2 promoter. (K-M) qRT-PCR and western blot assays were performed to assess the knockdown efficiency of siRNA specifically targeting TNK2. (N-R) CCK-8, wound healing, and transwell invasion assays were conducted to evaluate the effect of TNK2 knock-down on cell proliferation, migration, and invasion. Student’s t-test and one-way ANOVA were performed to analyze differences between groups. All data are presented as means ± standard deviations (SD). *P < 0.05, **P < 0.01, ***P < 0.001