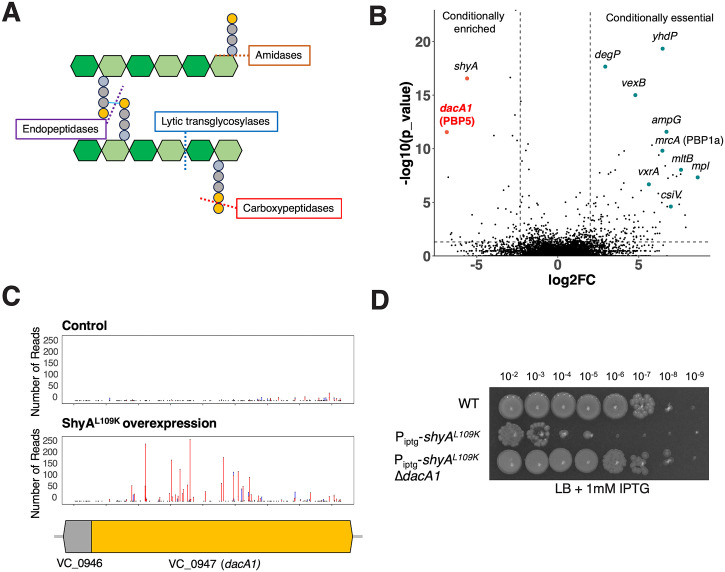
Fig 1. A screen for EP regulators identifies a genetic relationship between ShyA and PBP5.
(A) Schematic representation of autolysin cleavage sites in the cell wall. (B) Volcano plot of the change in relative abundance of insertion mutants between the control condition (WT) and the experimental condition (ShyAL109K overexpression) (X-axis) versus p-value (Y-axis). The dashed line indicates the cutoff criterion (>2-fold fitness change, P <0.005) for identification of genes that modulate ShyAL109K toxicity. (C) dacA1 was identified as conditionally enriched upon ShyAL109K overexpression using CON-ARTIST, red bars represent the number of reads from transposon insertions found in the forward orientation and blue bars represent insertions found in the reverse orientation. The black dots indicate every TA site available for transposon insertion (D) Overnight cultures of WT, Piptg-shyAL109K and Piptg-shyAL109K ΔdacA1 where plated on LB medium with 1mM IPTG and incubated overnight at 30°C.