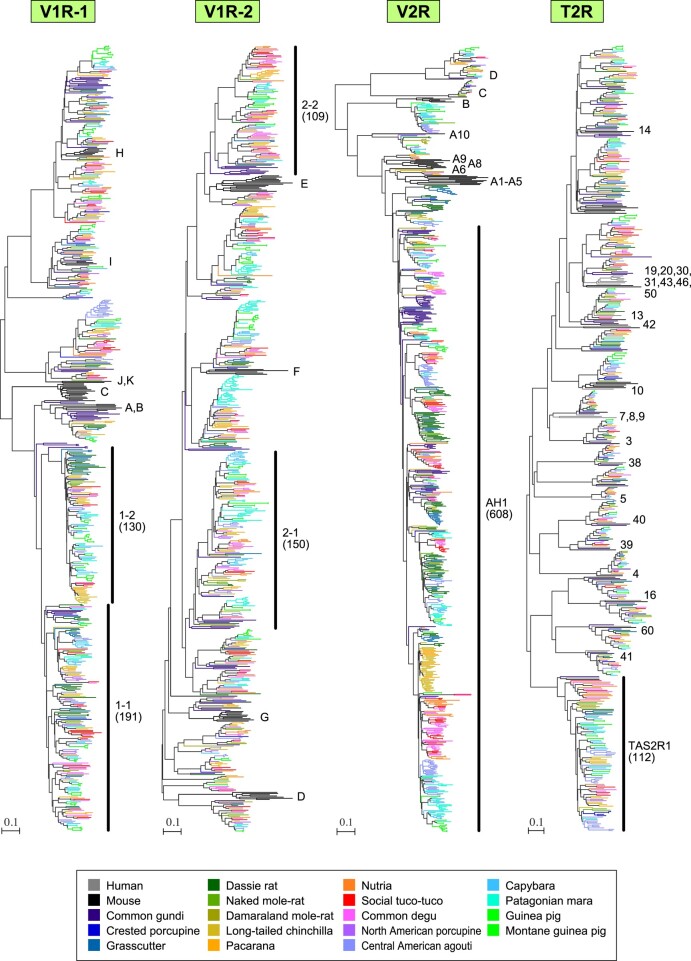
Fig. 4.
Phylogenetic trees for V1R, V2R, and T2R genes. The V1R genes were classified into two groups: V1R-1 containing clades A, B, C, H, I, J, and K and V1R-2 containing clades D, E, F, and G. V1R-1 and V1R-2 trees encompassed 613 and 624 V1R genes from 17 Hystricomorpha species, respectively, as well as 36 and 38 mouse V1R genes, respectively. The V2R tree was constructed using the exon 3 sequences of 761 hystricomorph V2R genes alongside 28 representative mouse genes. The T2R tree employed 517 hystricomorph T2R genes with 24 human and 36 mouse genes. The OGGs, which contained >100 genes from 17 Hystricomorpha species, are depicted by their OGG names along with the number of genes they include (in parentheses). For example, “1-1” in the V1R-1 tree denotes OGG-V1R-1-1, the largest OGG for V1R, housing 191 hystricomorph genes. In both V1R-1 and V1R-2 trees, the clade names for mouse genes align with those provided by Rodriguez et al. (2000) and Grus et al. (2005). In the V2R tree, subfamily names for mouse V2R genes are labeled according to Silvotti et al. (2011). In the T2R tree, gene names for human T2R genes are displayed (e.g. “14” represents TAS2R14). Color codes are outlined at the bottom, and the scale bar indicates the number of amino acid substitutions per site.