Abstract
Background
The major facilitator superfamily glucose transporters (GLUTs), encoded by solute carrier 2A (SLC2A) genes, mediate the transmembrane movement and uptake of glucose. To satisfy the improved energy demands, glycolysis flux is increased in cancers compared with healthy tissues. Multiple diseases, including cancer, have been associated with GLUTs. Nevertheless, not much research has been done on the functions of SLC2As in pan-cancer prognosis or their clinical treatment potential.
Methods
The SLC2A family genes' level of expression and prognostic values were analyzed in relation to pan-cancer. We then examined the association among SLC2As expression and TME, Stemness score, clinical characteristics, immune subtypes, and drug sensitivity. We merged bioinformatics analysis techniques with up-to-date public databases. Additionally, SLC2As from the KOBAS database were subjected to enrichment analysis.
Results
We discovered that SLC2As' gene expression differed significantly between normal tissues and many malignancies. A number of tumors from various databases demonstrate a relationship between prognosis and SLC2A family gene expression. For instance, SLC2A2 and SLC2A5 were associated with the overall survival (OS) of hepatocellular carcinoma. SLC2A1 was associated with the OS of lung adenocarcinoma and pancreatic adenocarcinoma. Moreover, the SLC2A family gene expression is significantly correlated with the pan-cancer stromal and immune scores, and the RNA and DNA stemness scores. Furthermore, we found that the majority of SLC2As had a strong correlation with the tumor stages in KIRC. The immunological subtypes and all members of the SLC2A gene family exhibited a substantial correlation. Moreover, pathways containing insulin resistance and adipocytokine signaling pathway may influence the progression of some cancers. Finally, there is a significant positive or negative connection between drug sensitivity and SLC2A1 expression.
Conclusion
Our research highlights the significant promise of SLC2As as prognostic indicators and offers insightful approaches for upcoming exploration of SLC2As as putative therapeutic targets in malignancies.
Keywords: SLC2A family genes, Pan-cancer, Prognosis, Tumor microenvironment, Therapeutic targets
1. Introduction
The Global Cancer Statistics 2020 report projects that there are expected to be 19.3 million new cases of cancer and about 10.0 million deaths from carcinoma-related causes in 2020 [1]. Furthermore, the global burden of cancer mortality and incidence is rising quickly. A genetic disorder known as cancer develops when a number of gene mutations that control cell division, survival, invasion, or other characteristics of the altered phenotype accumulate [2]. The development of targeted medicines and the discovery of driver mutations in tumors have been made possible by the tremendous influence of genomic analysis [3]. Nevertheless, there are still just a few tumors that contain targetable molecules, despite advancements in targeted therapy for particular cancer types. Furthermore, the efficacy of existing cancer therapy is restricted by tumor resistance to molecularly targeted medicines [4]. Therefore, in order to maximize combination therapy and raise these success rates, it is imperative to investigate additional unique possible molecular targets.
Tumor cells become hypoxic as a result of cancer progression, therefore hypoxia is a characteristic of tumors [5]. In order to survive, cancer cells must increase GLUT-1, cause apoptosis, and increase vascularization in response to the low pO2 [6]. The transition from glucose oxidative phosphorylation to "aerobic glycolysis," or the "Warburg effect," is one of the metabolic markers of tumors [7,8]. As a result, compared to normal cells, tumor cells require more glucose molecules. Particularly in the case of GLUT-3 and GLUT-1, enhanced expression of glucose transporters can significantly improve glucose absorption by cancer cells and generate energy via faster glycolysis [5,9]. The expression of the transporters GLUT-1 and GLUT-3 in cancer cells has been associated with factors that induce hypoxia (HIFs). Actually, GLUT-1 and GLUT-3 expression are induced by HIF activation [[10], [11], [12], [13]]. According to Pliszka et al., overexpression of GLUT1 or GLUT3 may be used as a marker of the aggressiveness of a cancer, its stage of carcinogenesis, the prognosis, and the overall survival (OS) of patients with certain tumors. They also proposed that the administration of glucose transporter inhibitors may be used in anticancer therapy [5].
The SLC2A genes encode the major facilitator superfamily glucose transporters (GLUTs), which are essential for mediating the transmembrane transit of different hexoses and their derivatives between diverse tissues [14,15]. Based on sequence similarity, the 14 GLUT proteins can be divided into 3 classes: Class 1 (GLUTs 1–4, 14); Class 2 (GLUTs 5, 7, 9, and 11); and Class 3 (GLUTs 6, 8, 10, 12, and HMIT) [15]. GLUT1–4 promote the uptake of glucose in numerous cell types, most notably erythrocytes, muscle and fat cells, endothelial cells of the blood–brain barrier, pancreatic β-cells, neurons, and multiple cancer cells [16]. Multiple diseases are related to SLC2As, including cancer, diabetes, and autoimmune illness [[17], [18], [19], [20], [21], [22], [23]]. Metabolic adaptation is intrinsic to tumorigenesis to satisfy the improved demands for biosynthetic, bioenergetic, and detoxification requirements of malignant cells [24]. Glycolysis flux is increased in cancers compared with healthy tissues [25]. For cell hyperproliferation, glucose absorption is a rate-limiting phase [14]. The overexpression of SLC2As and its functions in different malignancies have been shown in earlier research. This makes GLUT inhibition a possible option to treat cancer and decrease the growth of tumors [[26], [27], [28], [29], [30]]. Moreover, when paired with other treatments including radiation, immunotherapy, chemotherapy, targeted therapy, anti-metabolic medicines, or epigenetic medications, GLUTs inhibitors have demonstrated encouraging outcomes in the treatment of cancer [31]. SLC2A1-4 were studied best because of their role in cancer metabolism [29,32] and insulin resistance [[33], [34], [35], [36]]. The functional studies of SLC2A5-14 in human tumors are scarce and need to be extensively explored. Given their significant regulatory roles in various human cancers, a comprehensive understanding of SLC2As across different cancers is valuable and necessary to better elucidate their contributions to cancer development and potential therapeutic implications. Notably, more focus is being paid to how hypoxia promotes tumor immune evasion and immunosuppression [37]. Exploring the potential relationships between hypoxia and cancer is of great significance.
This study examined the baseline expression levels of SLC2As in normal tissues and human cancer samples from the TCGA database. The predictive significance of SLC2As in pan-cancer was then thoroughly assessed utilizing a variety of datasets, containing TCGA database and Kaplan-Meier Plotter. Additionally, the relationship among SLC2A family genes expression and TME, stemness score, and clinical stages were evaluated. Using enrichment analysis, we were able to anticipate potential pathways in which SLC2As may have contributed to carcinogenesis. We additionally evaluated at the connection between SLC2As expression and immunological subtype as well as medicine sensitivity in human malignancies. In total, SLC2As expression level and predictive value in pan-cancer were thoroughly assessed by our investigation, which also offered new information regarding SLC2As as possible therapeutic targets in different tumor types.
2. Materials and methods
2.1. Gene expression analysis
We utilized the UCSC Xena online database (https://xena.ucsc.edu/) to download the gene expression RNAseq (FPKM format), clinicopathological information, survival information, immunological subtype, and stemness score (DNA and RNA) of 33 tumors from the TCGA database [38]. The SLC2A family genes’ expression level was then extracted and integrated using the Perl software. Additionally, we investigated variations in SLC2A family gene expression among 33 distinct cancer groups using the "Wilcox test." The symbols "*", "**", and "***" denote P-values<0.05, <0.01, and <0.001, respectively. Moreover, in order to illustrate the expression profile of SLC2A family genes, the R packages “ggpubr” and “pheatmap” were used to draw a box plot and heatmap, respectively. The "corrplot" R-package was utilized to evaluate the relationship between the SLC2A family of genes.
2.2. Survival analysis
The Kaplan-Meier (KM) method and the log-rank test were used to explore the relationship between the expression of SLC2A family genes and the outcome of various malignancies in TCGA datasets. The R-package “survival” and “survminer” were applied for this research. Moreover, we performed a COX analysis to assess the relationship among SLC2As expression and outcome for all cancers. Lastly, we displayed the forest plot applying the R-package "forestplot" and "survival".
Additionally, in order to assess the association among the SLC2A family genes’ expression and prognosis in all tumors and their impact on clinical outcome (OS), the Kaplan-Meier Plotter online database (https://kmplot.com/analysis/) was consulted. The impact of the SLC2A family genes on the survival of patients with 21 distinct kinds of tumor was assessed using Kaplan-Meier Plotter database, which was derived from TCGA, gene expression omnibus (GEO), and European Genome-phenome Archive (EGA).
2.3. Immunohistochemistry data of HPA
The HPA immunohistochemistry data were utilized to confirm the SLC2A1, SLC2A2, and SLC2A5 protein expression levels in human normal tissues and malignancies. The expression of SLC2A1 (antibody CAB002759) was examined in lung cancer and normal lung tissues. Additionally, SLC2A1 protein expressions (antibody CAB002759) in normal pancreatic tissues and pancreatic cancer were examined. SLC2A2 (antibody HPA069490) and SLC2A5 (antibody HPA005449) protein expressions were examined in liver hepatocellular carcinoma and normal liver tissues.
2.4. Tumor immune microenvironment and stemness score analysis
The tumor microenvironment (TME) is made up of tumor cells, tumor stromal cells (stromal fibroblasts, endothelial cells, and immune cells like macrophages, lymphocytes, and microglia), as well as non-cellular extracellular matrix components like collagen, fibronectin, hyaluronan, and laminin, among others [39]. The stromal and immune cell scores of various tumors were determined applying the R-packages "estimate" and "limma" in conjunction with TCGA expression data. Using the Spearman's technique, the "corrplot" R-package was used to visualize the connection between the RNA stemness score (RNAss) and DNA stemness score (DNAss) and SLC2A family genes' expression. Additionally, the "limma", "reshape2″, "ggpubr", and "ggplot2″ packages were applied to do a relationship evaluation among SLC2A family genes' expression and TME and stemness score in particular malignancies.
2.5. Clinical characteristics analysis
The association among SLC2As and clinical characteristics was investigated in a subset of cancers. For this connection study, we chose the tumor stage. The association within the cancer stage and SLC2As was investigated using R-package "ggplot2″ in a subset of malignancies (BRCA, KICH, and KIRC) from the TCGA database.
2.6. Enrichment analysis
First, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis and Gene Ontology (GO) functional analysis were performed applying KOBAS database. Next, GO functional analysis and KEGG pathway analysis were performed using the Chinese bioinformatics website (http://www.bioinformatics.com.cn/).
2.7. Drug sensitivity and immune subtype analysis
The CellMiner™ database (https://discover.nci.nih.gov/cellminer/home.do) provided drug sensitivity processed data. Additionally, the R packages "ggplot2″, "limma", "impute", and "ggpubr" were utilized for processing the data and result visualization. Furthermore, the immunological subtype association study of SLC2As was performed employing the R-packages "reshape2″, "limma", and "ggplot2".
3. Results
3.1. The expression level of SLC2A family genes in pan-cancer
By analyzing these genes' expression in samples of human tumors and normal tissues, we hoped to learn more about the possible significance of SLC2A family genes in human malignancies. The TCGA database data showed that, in pan-cancer, SLC2A1, SLC2A3, SLC2A8, and SLC2A10 were highly expressed, SLC2A2, SLC2A7, and SLC2A14 were weakly expressed, while SLC2A4, SLC2A5, SLC2A6, SLC2A9, SLC2A11, SLC2A12, and SLC2A13 were moderately expressed (Fig. 1A). Additionally, the TCGA database was utilized to further assess SLC2A family genes expression and its connection in various forms of cancer. Findings indicated that SLC2A1 is most expressed in LUSC, SLC2A2, SLC2A3, SLC2A5, and SLC2A7 are most expressed in KIRC, SLC2A4, and SLC2A8 are most expressed in KICH, SLC2A6, and SLC2A11 are most expressed in CHOL, and SLC2A9, SLC2A10, SLC2A12, SLC2A13, and SLC2A14 are most expressed in HNSC, GBM, COAD, THCA, and KIRP, respectively (Fig. 1B). SLC2A2 and SLC2A9 showed the strongest positive correlation across all 18 cancer types, while SLC2A1 and SLC2A11 showed the exact opposite correlation (Fig. 1C).
Fig. 1.
SLC2A family gene expression levels and correlation in different cancer types from TCGA. (A) Boxplot illustrating the distribution of SLC2A family genes expression in various cancers. (B) Heatmap showing the difference of SLC2A family gene expression levels in different cancer types from TCGA database. The red and green indicate the high or low expression, respectively. (C) The correlation between SLC2A family genes. The blue dot indicated positive correlation and the red dot indicated negative correlation.
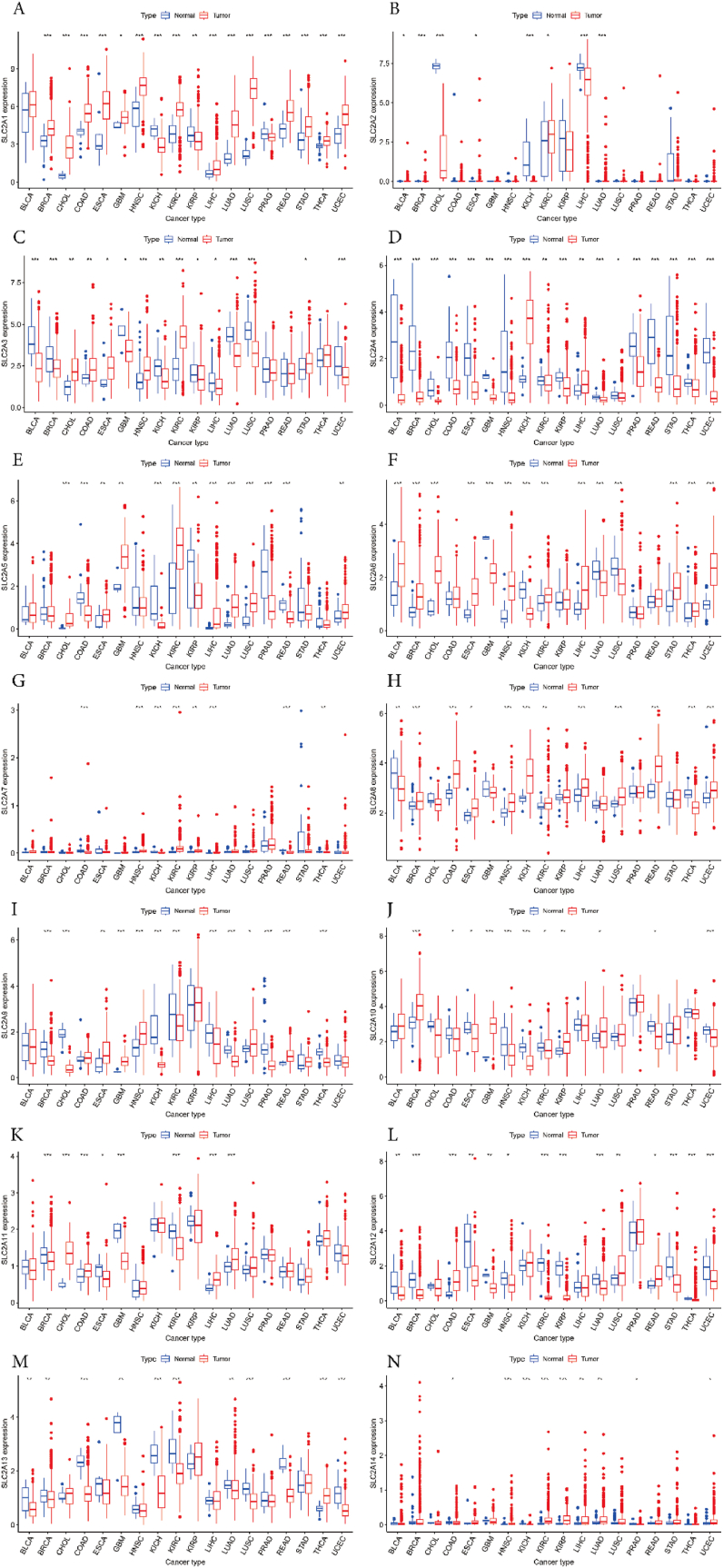
We next used RNA sequencing data from TCGA to assess the differential expressions of SLC2As in all tumors. Our findings found that SLC2A1 was higher expressed in breast invasive carcinoma (BRCA), cholangiocarcinoma (CHOL), colon adenocarcinoma (COAD), esophageal carcinoma (ESCA), head and neck squamous cell carcinoma (HNSC), kidney renal clear cell carcinoma (KIRC), liver hepatocellular carcinoma (LIHC), lung adenocarcinoma (LUAD), Lung squamous cell carcinoma (LUSC), rectum adenocarcinoma (READ), stomach adenocarcinoma (STAD), thyroid carcinoma (THCA), uterine corpus endometrial carcinoma (UCEC), and glioblastoma multiforme (GBM). Conversely, kidney renal papillary cell carcinoma (KIRP), Prostate adenocarcinoma (PRAD), and kidney chromophobe (KICH) were shown to have decreased expression (Fig. 2A). A higher expression of SLC2A2 was observed in a number of malignancies, including BRCA, LUAD, Bladder Urothelial Carcinoma (BLCA), ESCA, and KIRC. Meanwhile, lower expression was discovered in LIHC, KICH, and CHOL (Fig. 2B). SLC2A3 was found to be more highly expressed in HNSC, KIRC, CHOL, COAD, ESCA, and STAD. But in BLCA, BRCA, LUAD, LUSC, UCEC, KICH, GBM, KIRP, and LIHC, the level was lower (Fig. 2C). In addition, SLC2A4 was more expressed in KICH, and LIHC. The lower expression was in BLCA, BRCA, CHOL, COAD, ESCA, GBM, HNSC, KIRP, LUAD, PRAD, READ, STAD, THCA, UCEC, KIRC, and LUSC (Fig. 2D). In CHOL, KIRC, LIHC, LUAD, LUSC, ECSA, GBM, and UCEC, SLC2A5 expression was greater. However, SLC2A5 expression was reduced in COAD, KICH, PRAD, READ, and KIRP (Fig. 2E). Additionally, we found that SLC2A6 had greater expression levels in a number of malignancies, including BLCA, BRCA, CHOL, ESCA, HNSC, KIRC, LIHC, STAD, THCA, and UCEC. But SLC2A6 expression was shown to be decreased in GBM, KICH, LUAD, and LUSC (Fig. 2F). Moreover, analyzing revealed that SLC2A7 expression was higher in HNSC, KIRC, LIHC, and KIRP. SLC2A7’ expression was decreased in COAD, KICH, READ, and THCA (Fig. 2G). Further investigation revealed that SLC2A8 is overexpressed in a number of malignancies as compared to normal tissues, including BRCA, COAD, HNSC, KICH, LIHC, LUSC, READ, UCEC, KIRC, and ESCA. On the other hand, SLC2A8 expression decreased in BLCA and THCA (Fig. 2H). Furthermore, it was found that GBM, HNSC, READ, ESCA, and LUSC had greater levels of SLC2A9 expression. The lower SLC2A9 expression level was found in BRCA, CHOL, KICH, KIRC, LIHC, LUAD, PRAD, and THCA (Fig. 2I). Furthermore, we observed that SLC2A10 was upregulated in KIRP, LUAD, GBM, and BRCA. Conversely, HNSC, KICH, UCEC, COAD, ESCA, KIRC, and READ were shown to have reduced SLC2A10 levels (Fig. 2J). The expression of SLC2A11 was higher in CHOL, COAD, LIHC, and LUAD. Concurrently, BRCA, GBM, KIRC, and ESCA have decreased expression of SLC2A11 (Fig. 2K). SLC2A12 was expressed more in COAD, LUSC, and READ. Meanwhile, a lower SLC2A12 expression was discovered in BRCA, KIRC, KIRP, LUAD, STAD, THCA, UCEC, BLCA, ESCA, GBM, and HNSC (Fig. 2L). Besides, a higher SLC2A13 expression was found only in THCA. But in COAD, KICH, KIRC, LUSC, READ, UCEC, BLCA, BRCA, GBM, and LUAD, there was less expression (Fig. 2M). SLC2A14 was found to be more highly expressed in HNSC, KIRC, KIRP, LIHC, LUAD, and UCEC. In KICH, COAD, and PRAD, the lower expression was present concurrently (Fig. 2N).
Fig. 2.
SLC2A family gene expression levels in different cancer types and normal tissue. (A) SLC2A1 (B) SLC2A2 (C) SLC2A3 (D) SLC2A4 (E) SLC2A5 (F) SLC2A6 (G) SLC2A7 (H) SLC2A8 (I) SLC2A9 (J) SLC2A10 (K) SLC2A11 (L) SLC2A12 (M) SLC2A13 (N) SLC2A14. The blue boxplots indicate the normal tissues and the red boxplots indicate the cancer tissues. *P < 0.05, **P < 0.01, and ***P < 0.001.
3.2. Prognostic value of SLC2A family genes in pan-cancer
In order to look into the connection between prognostic value and the expression level of the SLC2A family genes, we employed various databases. Notably, Kaplan-Meier survival curves demonstrated that the expression of SLC2A family genes was linked with prognosis in various kinds of cancer (Fig. 3, S1). SLC2A1 proved useful throughout the KIRP procedure. On the other hand, SLC2A1 negatively impacted ACC, LUAD, LIHC, SARC, PAAD, MESO, and SKCM (Fig. 3A–S1A). In addition, SLC2A2 acted as a buffer in both KIRC and LIHC (Fig. 3B–S1B). SLC2A3 was a high-hazard feature in STAD, UVM, HNSC, MESO, LGG, and GBM (Fig. 3C–S1D). Additionally, the data demonstrated a correlation between prolonged survival periods and increased SLC2A4 expression in patients with LGG, MESO, LAML, and DLBC. While, SLC2A4 had a detrimental significance in UCEC (Fig. 3D–S1E). In three distinct malignancies that contained SKCM, SARC, and THCA, SLC2A5 acted as a protective factor. On the other hand, SLC2A5 was deleterious to the development of LAML, KICH, and LIHC malignancies (Fig. 3E–S1F). SLC2A6 was found to be a protective gene in UCEC, BLCA, and ESCA. SLC2A6 was found to be a protective gene in ESCA, BLCA, and UCEC. Nevertheless, SLC2A6 was a risky gene in KIRC, UVM, MESO, COAD, ACC, and BLCA (Fig. 3F–S1G). The five cancer types that included UCEC, LUAD, OV, ACC, and LGG were all negatively impacted by SLC2A7 (Fig. 3G–S1H). Besides, SLC2A8 had a protective effect on four different types of tumors, containing CESC, PAAD, BLCA, and HNSC. Conversely, in LAML, SLC2A8 was a high-risk gene (Fig. 3H–S1I). For UCS, KIRP, and KIRC, SLC2A9 was a low-risk gene. SLC2A9 participated in LGG, LAML, and CESC concurrently as a high-risk factor (Fig. 3I–S1J). Additionally, we discovered that SLC2A10 had a protective effect on PCPG and MESO. However, SLC2A10 posed a risk in LGG, UVM, and PAAD (Fig. 3J–S1K). Additionally, our findings demonstrated the protective role of SLC2A11 in PAAD, MESO, LUAD, LGG, and KIRP (Fig. 3K, S1L). SLC2A12 was a beneficial factor in KIRP and LAML. In contrast, SLC2A12 was damaging to UCEC and BLCA (Fig. S1M). SLC2A13 was a protective gene in KIRC and LGG (Fig. 3L). Moreover, SLC2A14 played a beneficial role in READ. However, SLC2A14 was linked to a significant risk of LGG and ACC (Fig. S1C). Furthermore, we found that SLC2A4/11/13 provided protection against LGG. In contrast, SLC2A3/7/9/10/14 were risk factors for LGG. Regarding MESO patients, SLC2A4/10/11 were considered low-risk variables. Conversely, poorer MESO results were associated with elevated SLC2A1/3/6 expressions.
Fig. 3.
Survival analysis of SLC2A family genes across multiple cancer types. (A) SLC2A1 (B) SLC2A2 (C) SLC2A3 (D) SLC2A4 (E) SLC2A5 (F) SLC2A6 (G) SLC2A7 (H) SLC2A8 (I) SLC2A9 (J) SLC2A10 (K) SLC2A11 (L) SLC2A13. The red line in the photos indicates high expression and the blue line in the photos indicates low expression. P value less than 0.05 is considered as difference.
Using COX analysis, we examined the prognostic significance of SLC2As in all cancers (Fig. S2). Our results shown that SLC2A1 played a protective prognostic factor in KICH, THYM, ACC, LIHC, KIRP, MESO, LUAD, SARC, PAAD, UCEC, CESC, and SKCM. On the other hand, LIHC and KIRC, SLC2A2 was a poor predictive predictor. The low-risk gene in THCA, ACC, UVM, MESO, CESC, LGG, HNSC, STAD, GBM, BLCA, PAAD, and KIRC was SLC2A3. SLC2A4 played protective roles in CHOL and READ. On the other hand, a higher incidence of LGG and LIHC was linked to SLC2A4. For KICH, LAML, and LIHC, SLC2A5 was a low-risk factor. SLC2A5 also has a negative role in SKCM and SARC. SLC2A6 functioned as a safeguard prognosis marker in KIRC, UVM, ACC, COAD, MESO, and LIHC. On the other hand, SLC2A6 was the dangerous gene in LGG and SARC. SLC2A7 was a protecting gene in LGG, MESO, BRCA, OV, and UCEC. SLC2A8 exhibited a protective effect in THYM and LAML. SLC2A8 served as a hazardous factor in PAAD, CESC, and BLCA in the meantime. A higher prognosis was linked to higher levels of SLC2A9 expression in patients with LGG, THCA, and HNSC. Conversely, a vulnerable gene in both KIRP and KIRC was SLC2A9. Additionally, according to our findings, SLC2A10 provided protection against LGG, UVM, PAAD, MESO, and GBM. Contrarily, SLC2A10 was a high-risk prognosis factor in PCPG. SLC2A11 was the low-risk gene in DLBC. While SLC2A11 served as a harmful gene in PAAD, MESO, KIRP, LGG, LUAD, and OV. SLC2A12 functioned a harmless component in UCEC and BLCA. Besides, SLC2A13 provided protection in MESO. However, SLC2A13 was a harmful gene in LGG, KIRC, and SKCM. SLC2A14 was a beneficial gene in BLCA, LGG, CESC, ACC, and THCA. These findings suggested that SLC2As may have varying effects on prognostic significance in various cancer types.
Next, using data from the TCGA, GEO, and EGA databases, the Kaplan-Meier plotter was utilized to evaluate the survival (OS) connected to the SLC2A gene. SLC2A2/7 had negative impacts on BRCA, as Fig. 4A illustrates. Comparable outcomes are also observed in OV and UCEC (Fig. 4G–J). Furthermore, in CESC, SLC2A8 played a protective predictive role (Fig. 4B). In KIRP, SLC2A1 had a negative predictive effect. SLC2A9/11 functioned as a low-risk factor in KIRP concurrently (Fig. 4C). SLC2A2/9/13 showed protective effects on KIRC, while SLC2A1/3/6 played harmful prognostic roles in KIRC (Fig. 4D). A dangerous gene in both LUAD and LIHC was SLC2A1. Conversely, SLC2A2 and SLC2A11 provided protection in LIHC and LUAD, respectively (Fig. 4E and F). SLC2A1 was deleterious to PAAD, while SLC2A8/11 was low-risk (Fig. 4H). Furthermore, SLC2A3 posed a risk for STAD (Fig. 4I).
Fig. 4.
Overall survival curves comparing the high and low expression of SLC2A family genes in various cancers from Kaplan-Meier Plotter database.
Nevertheless, the results above from several databases revealed some inconsistent information about the SLC2A family genes’ expression in particular malignancies (Table S1). These contradicting results were caused by diverse data gathering methods and putative processes with varying biological properties. Additionally, it was discovered that SLC2A1 expression might function as a separate OS prognostic biomarker in lung and pancreatic adenocarcinomas by comparing and evaluating the prognostic significance of SLC2As in cancers in various databases. Furthermore, we discovered that the expression of SLC2A2 and SLC2A5 may function as separate prognostic indicators for OS in hepatocellular carcinoma. Moreover, SLC2A1 protein expression in lung and pancreatic adenocarcinomas, as well as SLC2A2 and SLC2A5 protein expression in hepatocellular carcinoma, were confirmed by immunohistochemical data (Fig. S3).
3.3. Correlation among SLC2A family genes expression and TME, stemness score in pan-cancer
Since our outcomes have verified the prognostic relevance of SLC2A family gene in all cancers, it is highly suitable to investigate additional correlations between the expression of SLC2As and TME in all cancers. The results of the TME related scores showed that the majority of SLC2As expression was strongly inversely correlated with tumor purity and considerably positively correlated with stromal score, immune score, and estimate score (Fig. 5A–D). Our findings indicate that there seems to be a consistent positive association between SLC2A3/5/6/9 and the stromal score, immune score, and estimate score in all malignancies. At the same time, SLC2A11's unfavorable connection with these scores was constant for all malignancies. Furthermore, SLC2As and immune score in LGG, TGCT, THCA, and THYM were revealed to be clearly correlated. Likewise, there was a strong positive or negative connection seen in pan-cancer among the expression of SLC2A family genes and DNAss and RNAss (Fig. 5E and F). For instance, SLC2A9/12 expression was inversely associated with DNAss in OV. SLC2A1, on the other hand, had a favorable relationship with DNAss in OV. Moreover, we found that SLC2A2/12 and DNAss in TGCT had a statistically significant negative connection. At the same time, SLC2A14 had a positive correlation with DNAss in TGCT (Fig. 5E). We found that SLC2A4/10/13 had a negative relationship with RNAss in THYM during the RNAss study. Additionally, SLC2A3 strongly correlated negatively with RNAss in a number of malignancies, including THCA, GBM, BLCA, and KICH (Fig. 5F). Thus, our findings suggested that the ability of genes belonging to the SLC2A family to control the immune microenvironment varies amongst malignancies.
Fig. 5.
Correlation of SLC2A family genes expression with tumor microenvironment, Stemness score in pan-cancer. The SLC2A family genes associated Stromal score (A), Immune score (B), ESTIMATE score (C), Tumor purity (D), DNAss (E), and RNAss (F) are illustrated. Red dots indicate a positive correlation, and blue dots indicate a negative correlation.
3.4. Association between SLC2A family genes expression and TME, stemness score in selected cancers
Using gene expression analysis, it was discovered that, while the patterns varied by gene, all SLC2A family genes were expressed differently in KIRC compared to normal tissues. Additionally, only SLC2A4 and SLC2A8 were up-regulated whereas the other genes were down-regulated in KICH, suggesting differential expression of the remaining SLC2A family genes, with the exception of SLC2A11 and SLC2A12. Similarly, other SLC2A family genes are expressed differently in BRCA compared to normal tissues, with the exception of SLC2A5/7/14. Consequently, we used information from the database of the TCGA to explore the connection among the expression of SLC2A family genes and TME and Stemness scores in these three kinds of tumors. SLC2A1/5/6/14 were favorably correlated with RNAss in BRCA, while SLC2A3/4/7/8/9/10/11/12/13 were negatively correlated with RNAss, as Fig. 6A illustrates. Additionally, in BRCA, SLC2A1/5/6/14 had a positive correlation with DNAss and SLC2A9/10 had a negative correlation with DNAss. We found that SLC2A2/3/4/5/6/7/9/10/12/13 had a positive correlation with stromal score during the TME analysis, whereas SLC2A1/8/11 and BRCA's stromal score showed a negative link. Furthermore, SLC2A1/10/11 had an adverse connection with immunological score in BRCA, but SLC2A3/4/5/6/7/9/12/14 had a positive correlation. The BRCA estimate score was favorably correlated with the expression of SLC2A3/4/5/6/7/9/12/13/14. In addition, there was a negative correlation between SLC2A1/10/11 and the estimate score in BRCA.
Fig. 6.
Correlation analysis of SLC2A family genes expression with Stemness score and tumor microenvironment in three cancer types. (A) BRCA (B) KICH (C) KIRC. R means correlation value, positive number means positive correlation, negative number means negative correlation.
Additionally, in KICH, SLC2A4 exhibited a positive relationship with RNAss while SLC2A3/5/6 had a negative relationship with RNAss. DNAss in KICH was adversely correlated with SLC2A5/6. According to the findings of the TME correlation study, SLC2A3/5/6/10 showed a positive association with the stromal score in KICH, while SLC2A11 had a negative correlation. The immune score in KICH was strongly correlated with SLC2A3/5/6. In KICH, the expression of SLC2A3/5/6/10 was positively connected with the estimate score, whereas SLC2A4/11 expression had a negative correlation (Fig. 6B).
SLC2A1/3/4/6/10 and SLC2A2/11/13 had negative correlations and positive correlations with RNAss in KIRC, respectively, as Fig. 6C illustrates. In addition, SLC2A2/5/7/9/13 expression was inversely correlated with DNAss in KIRC. TME analysis showed that SLC2A1/3/6/10 and the stromal score correlated positively with KIRC, but negatively with SLC2A2/4/8/11/12. In addition, SLC2A5/6/14 expression and immune score in KIRC were positively connected, while SLC2A4/11/12 was negatively correlated. SLC2A3/5/6/10/14 showed a favorable correlation with the KIRC estimate score. Concurrently, there was a negative association between the estimate score in KIRC and SLC2A2/4/11.
3.5. Correlation among clinical characteristics and SLC2A family genes expression in selected tumors
Three cancer types (BRCA, KICH, and KIRC) were chosen for SLC2A family gene expression levels and tumor stage association study based on TCGA database gene expression analysis (Fig. S4). We discovered that in BRCA, only the expression of SLC2A1 and SLC2A4 correlated with tumor stage (Fig. S4A). Additionally, we discovered that the expression of other SLC2A family genes in KICH did not significantly correlate with tumor stage, with the exception of SLC2A5 (Fig. S4B). While, most of SLC2As were significantly associated with tumor staging in KIRC, including SLC2A4/6/9/10/11/13/14 (Fig. S4C). However, different genes are expressed differently in KIRC. For example, SLC2A4/9/10/13 expression was markedly upregulated in KIRC tumors at an early stage. On the other hand, SLC2A6/14 expression dramatically increased in KIRC tumors that were advanced in stage.
3.6. Correlation among SLC2A family genes expression and immune subtype
We also looked into the relationship of the expression of the SLC2A family genes and the immune subtype in pan-cancer. Our findings demonstrated a significant relationship among the expression of all SLC2A gene family members and immune subtypes C1 (wound healing), C2 (IFN-γ dominant), C3 (inflammatory), C4 (lymphocyte deficient), C5 (immunologically silent), and C6 (TGF-β dominant) in all tumors [38,40] (Fig. 7A). Next, we chose three types of tumors to study including BRCA, KICH, and KIRC. Significant relationships were found between SLC2A1/3/4/5/6/9/10/11/12/13/14 and the immunological subtype in BRCA (Fig. 7B). For instance, SLC2A4 and SLC2A13 expressed more in C3 and C6, while SLC2A3/5/9 expression was up-regulated in C6. Fig. 7C demonstrated the correlation between the immunological subtype in KICH and the expressions of SLC2A3/5/6/9/10. Also, these genes overexpressed in C1, with the exception of SLC2A9. Moreover, we discovered that there were substantial differences between SLC2A1/2/3/4/5/6/7/9/11/12/13/14 in KIRC (Fig. 7D). C3 had greater levels of SLC2A2/9/13 expression. On the other hand, C5 had increased SLC2A4/11/12 expression. The selectivity of SLC2A members in various cancers was demonstrated by these data once more.
Fig. 7.
SLC2A family genes expression levels of different immune subtype in pan-cancer, and specific three cancer types. (A) Pan-cancer (B) BRCA (C) KICH (D) KIRC. X axis represents immune subtype, Y axis represents gene expression. C1, wound healing; C2, IFN-γ dominant; C3, inflammatory; C4, lymphocyte depleted; C5, immunologically quiet; C6, TGF-β dominant. *P < 0.05, **P < 0.01, and ***P < 0.001.
3.7. Functional and pathway analyses of SLC2A family genes in human tumors
Next, using the KOBAS database, we investigated GO functions and KEGG pathways connected to SLC2A family genes in human malignancies. Seven enriched pathways and the expression of SLC2A family genes were shown to be strongly correlated by the KEGG enrichment analysis. These pathways included insulin secretion, insulin resistance, the glucagon signaling pathway, the adipocytokine signaling pathway, central carbon metabolism in cancer, type II diabetes mellitus, and the digestion and absorption of carbohydrates (Fig. S5A). Furthermore, the KOBAS database's GO enrichment analysis revealed every cell biological function. SLC2A family genes significantly related to glucose transmembrane transport (Fig. S5B). In addition, KEGG illness analysis revealed that ldiopathic generalized epilepsies, hereditary stomatocytosis, and cardiovascular diseases were the top three disorders associated with the expression of SLC2A family genes (Fig. S5C).
3.8. Drug sensitivity analysis
The comprehensive analysis was carried out with the goal of investigating any possible relationship among sensitivity to drugs and SLC2As expression in different human tumor cell lines from the CellMiner™ database. The outcome showed a favorable relationship among SLC2A1 expression and medicine sensitivity of Kahalide F (Fig. 8B), ENMD-2076 (Precursor) (angiogenic kinase inhibitor) (Fig. 8D and E), Staurosporine (Fig. 8H), CCT-128930 (CCT-128930) (Fig. 8I), JNJ-38877605 (c-Met inhibitor) (Fig. 8K), VE-821 (the ATR inhibitor) (Fig. 8M), AZD-5363 (cell cycle inhibitor) (Fig. 8N), SGX-523 (c-Met inhibitor) (Fig. 8O), and Saracatinib (Fig. 8P). Furthermore, Fig. 8 illustrates the substantial inverse relationship between SLC2A1 expression and the drug sensitivity of Lapachone (Fig. 8A), tic10 (also known as ONC-201, TRAIL-inducing compound) (Fig. 8C), Hydrastinine HCI (Fig. 8F), Imexon (Fig. 8G), Cpd-401 (Fig. 8J), and Amuvatinib (Fig. 8L). The findings could have an impact on clinical medicine because of the SLC2A family genes' drug sensitivity.
Fig. 8.
Drug sensitivity analysis of SLC2A family genes. (A) Lapachone (B) Kahalide F (C) tic10 (D) ENMD-2076 (E) ENMD-2076 Precursor (F) Hydrastinine HCI (G) Imexon (H) Staurosporine (I) CCT-128930 (J) Cpd-401 (K) JNJ-38877605 (L) Amuvatinib (M) VE-821 (N) AZD-5363 (O) SGX-523 (P) Saracatinib. Cor, correlation coefficient.
4. Discussion
SLC2As may be crucial factors in malignancies and may have therapeutic applications, according to increasing evidence. This work was the first to determine the basic expression levels of SLC2A family genes in 33 types of cancer and in pericancerous or normal tissues from TCGA database. According to our findings, SLC2A1 expression was downregulated in three tumor types and highly upregulated in fourteen cancer types as compared to normal tissues. However, eight distinct malignancies were linked to aberrant SLC2A2 expression, three of which were down-regulated and five of which were up-regulated. Of the six cancer types, SLC2A3 expression was higher in six whereas it was lower in nine. Furthermore, in comparison to normal tissues, SLC2A4 was downregulated in sixteen cancer types and upregulated in two cancer types. Eight cancer types had higher levels of SLC2A5 expression, while five cancer types had lower levels of the gene. Ten different cancer types had higher expression levels of SLC2A6 and SLC2A8, whereas four and two cancer types, respectively, had lower expression levels of these two proteins. Furthermore, compared to normal tissues, SLC2A7 and SLC2A11 were expressed more in four cancer types and less in four cancer types. Thirteen distinct malignancies, eight of which were down-regulated and five of which were up-regulated, were linked to aberrant SLC2A9 expression. SLC2A10 was up-regulated in four types of cancers and lower-regulated in seven tumor kinds. Eleven kinds of cancer had fewer expression levels of SLC2A12, whereas three cancer kinds had greater expression levels. Besides, we discovered that SLC2A13 was higher-regulated in one tumor type, while SLC2A13 was lower expressed in ten cancer types. Three cancer types showed down-regulation of SLC2A14 expression, whereas six cancer types showed up-regulation of the gene. Next, we compiled all significant survival-related data from several databases. The SLC2As have a strong correlation with cancer patient survival. For instance, SLC2A1 was found to be deleterious in LIHC, LUAD, PAAD, and SARC, this finding was similar across many databases and the majority of prior research [[41], [42], [43], [44], [45], [46], [47], [48], [49]]. Also, we discovered that in LIHC and KIRC, improved OS was correlated with increased expression of SLC2A2. Hye et al. indicated that SLC2A2 is a promising target for hepatocellular carcinoma [50]. Yun et al. also found that SLC2A2 was associated with clinical stages and independently related to OS in patients with HCC [51], which was consistent with our findings. Besides, we also observed that SLC2A5 was a detrimental factor in LIHC. And Song et al. constructed a prognostic signature contained SLC2A5, which can help to predict the OS of HCC and explore the potential therapeutic targets of HCC [52]. Overall, our findings clearly show that SLC2A family genes are useful prognostic indicators for all cancer types.
Growing research has recently suggested that TME may have significant impacts on immune evasion, tumor metastasis, micro angiogenesis, and proliferation [[53], [54], [55]]. Our findings showed a link between TME in all tumors and SLC2A family genes expression in all tumors. Especially, SLC2A3/5/6/9 expression in the stromal score, immune score, and estimate score appeared to have a constant positive connection in all malignancies, indicating that SLC2A3/5/6/9 are expected to be effective immunomodulatory variables. Furthermore, there is a substantial association between the stromal and immune factors in TME and BRCA, KICH, and KIRC. These findings suggest that SLC2A family genes may be valuable treatment targets in certain malignancies, although further correlation research is required to confirm this. Shin et al. came to the conclusion that stromal cells and breast cancer cells engage in metabolic interactions, and the reverse Warburg effect may indicate glucose metabolism in tumor stromal cells. The reverse Warburg effect suggests that cancer-associated fibroblasts (CAFs), which are found in the breast cancer stroma, are the site of aerobic glycolysis. Additionally, GLUT1 expression was higher in breast CAFs than in normal fibroblasts, and CAFs had higher levels of glycolysis and genes associated to glucose transporters in the co-culture investigation of breast cancer cells and fibroblasts [56]. However, there are few studies on the correlation between TME and SLC2A family genes in KICH and KIRC, which can be an important direction in the future. Additionally, we discovered a positive or negative association between the pan-cancer stemness score and the genes of the SLC2A family. This examination of the association between DNAss and RNAss and the SLC2A family genes is thought to be helpful for patients responding to stem cell-based immunotherapy.
Moreover, in a subset of malignancies, the relationship between SLC2As expression and tumor stages was examined. Moreover, we found that the majority of SLC2As had a strong correlation with the tumor stages in KIRC, which led us to consider SLC2As' potential applications as markers for prognosis and diagnosis in KIRC. Mariana et al. observed that, in comparison to patients with III−IV cancer stages, RCC patients with I−II cancer stages had a considerably greater level of hsa-miR-144-5p (hsa-miR-144-5p has been shown to directly regulate GLUT-1) [57]. However, our results do not show that SLC2A1 is associated with the tumor stages of KIRC. Therefore, a great deal more research is required to confirm in the future the relationship between SLC2A1 and tumor stages in KIRC. Additionally, the possible relationship among the SLC2A family genes’ expression and immune subtypes was examined in both all cancers and specific tumor types (BRCA, KICH, and KIRC). According to our findings, the majority of SLC2As had strong relationships with BRCA, KICH, and KIRC, and all of them were substantially correlated with immune subtypes in pan-cancer.
Enrichment analysis was employed to explore the roles of SLC2As in cancer progression. We found that SLC2As were related to many diseases, including cardiovascular diseases, hereditary stomatocytosis, and ldiopathic generalized epilepsies. In addition, our results also showed that SLC2As significantly correlated with glucose transmembrane transport activity, which indicated that SLC2As may be associated with cancer development. The KEGG analysis presented here indicated that SLC2As are involved in these correlation pathways. The association between insulin-resistant disorders, like obesity and type 2 diabetes, and cancers of the colon, liver, pancreas, breast, and endometrium was among the numerous epidemiological and clinical data that supported this theory [58]. Additionally, via participating in the adipocytokine signaling pathway, SLC2As may affect the development and malignant evolution of breast cancer [59,60]. In summary, further research on the interaction between SLC2As and targeted drugs may benefit from an understanding of these pathways.
Based on previous studies and our analysis, we observed that SLC2As can be served as predictive indicators for several types of cancer, specially, the prognosis value of SLC2A1 in LIHC, LUAD, PAAD, and SARC, SLC2A2 and SLC2A5 in LIHC. On the one hand, we need to continuously increase the prognostic value analysis of SLC2A1/2/5 genes in these tumors. On the other hand, we can also try the survival analysis and prognostic study of other SLC2A family genes in cancers in the future. Furthermore, diagnostic utility of SLC2A family genes in KIRC may present a viable path forward. Above all, we think that SLC2As may be useful in anticipating the response to immunotherapy and could be a novel immunotherapy target for a variety of tumors because of the strong link between SLC2As and TME as well as the findings of other studies. Consequently, our findings offer novel perspectives for further clinical research and tumor immunotherapy, as well as novel directions for the investigation of SLC2A inhibition as a possible cancer therapeutic approach.
Genes encoding phosphoglucose isomerase (PG), glucose transporters, hexokinase 2 (HK2), HIF-1, and glyceraldehyde 3-phosphate dehydrogenase are all altered by tumor cells and contribute to the hypoxic environment found in malignancies. Studies have indicated that hypoxia-regulated genes may be targets for therapeutic intervention in the treatment of cancer [5]. As previously noted, higher expression of GLUT1 and GLUT3 is a result of a hypoxic state. Additionally, our findings revealed that cancer cells overexpress a number of GLUTs, most notably GLUT1. Furthermore, GLUT1 overexpression may function as a prognostic biomarker for a variety of cancer types, such as SARC, LUAD, LIHC, and PAAD. Furthermore, as a novel target in anticancer therapy, GLUT1 can be addressed. Our findings thus suggest an indirect connection between hypoxia and malignancy. The latest work may assist direct targeted therapy for many tumors and offer fresh insights into how hypoxia influences cancer prognosis.
Nevertheless, our study has a number of shortcomings. Firstly, as our research's conclusions on SLC2As were primarily derived from the computational analysis of genetic data, additional in vivo and in vitro tests are required to confirm our findings. Furthermore, the minimal correlation we found in our study between SLC2As and immunotherapy meant that further experimental research and clinical trials would be necessary to confirm the findings.
5. Conclusion
In summary, the current study demonstrated the intricate and wide-ranging functions that the expression of SLC2A family genes plays in cancer development and medical results, suggesting that SLC2As may be useful prognostic biomarkers for particular tumor types. The SLC2As were linked to immune subtypes, stemness score, and TME, which offers novel prospects for immunotherapy research and fresh perspectives on the SLC2A family genes as possible targets in malignancies. The association between hypoxia and malignancies is further clarified by our study, which also offers new evidence supporting SLC2A1 as a potential therapeutic target for a range of cancers.
Funding
Not applicable.
Data availability statement
Data included in article/supplementary material/referenced in article.
CRediT authorship contribution statement
Yating Liu: Writing – original draft, Conceptualization. Xinyu Li: Data curation. Jie Yang: Data curation. Shanshan Chen: Formal analysis. Changyu Zhu: Investigation. Yijun Shi: Software. Shoutao Dang: Software. Weitao Zhang: Validation. Wei Li: Writing – review & editing, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Not applicable.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e29655.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., et al. Global cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries [J] CA Cancer J Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Chakravarty D., Solit D.B. Clinical cancer genomic profiling [J] Nat. Rev. Genet. 2021;22(8):483–501. doi: 10.1038/s41576-021-00338-8. [DOI] [PubMed] [Google Scholar]
- 3.Mani D.R., Krug K., Zhang B., Satpathy S., Clauser K.R., Ding L., et al. Cancer proteogenomics: current impact and future prospects [J] Nat. Rev. Cancer. 2022;22(5):298–313. doi: 10.1038/s41568-022-00446-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holohan C., VAN Schaeybroeck S., Longley D.B., Johnston P.G. Cancer drug resistance: an evolving paradigm [J] Nat. Rev. Cancer. 2013;13(10):714–726. doi: 10.1038/nrc3599. [DOI] [PubMed] [Google Scholar]
- 5.Pliszka M., Szablewski L. Glucose transporters as a target for anticancer therapy [J] Cancers. 2021;13(16) doi: 10.3390/cancers13164184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mckenzie S., Sakamoto S., Kyprianou N. Maspin modulates prostate cancer cell apoptotic and angiogenic response to hypoxia via targeting AKT [J] Oncogene. 2008;27(57):7171–7179. doi: 10.1038/onc.2008.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Warburg O. The chemical constitution of respiration ferment [J] Science. 1928;68(1767):437–443. doi: 10.1126/science.68.1767.437. [DOI] [PubMed] [Google Scholar]
- 8.Warburg O. On the origin of cancer cells [J] Science. 1956;123(3191):309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 9.Sampedro-NúñEZ M., Bouthelier A., Serrano-Somavilla A., MartíNEZ-HernáNDEZ R., Adrados M., MartíN-PéREZ E., et al. LAT-1 and GLUT-1 carrier expression and its prognostic value in gastroenteropancreatic neuroendocrine tumors [J] Cancers. 2020;12(10) doi: 10.3390/cancers12102968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koh Y.W., Lee S.J., Park S.Y. Differential expression and prognostic significance of GLUT1 according to histologic type of non-small-cell lung cancer and its association with volume-dependent parameters [J] Lung Cancer. 2017;104:31–37. doi: 10.1016/j.lungcan.2016.12.003. [DOI] [PubMed] [Google Scholar]
- 11.Mazumder S., Higgins P.J., Samarakoon R. Downstream targets of VHL/HIF-α signaling in renal clear cell carcinoma progression: mechanisms and therapeutic relevance [J] Cancers. 2023;15(4) doi: 10.3390/cancers15041316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beylerli O., Sufianova G., Shumadalova A., Zhang D., Gareev I. MicroRNAs-mediated regulation of glucose transporter (GLUT) expression in glioblastoma [J] Noncoding RNA Res. 2022;7(4):205–211. doi: 10.1016/j.ncrna.2022.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blaszczak W., Barczak W., Masternak J., Kopczyński P., Zhitkovich A., Rubiś B. Vitamin C as a modulator of the response to cancer therapy [J] Molecules. 2019;24(3) doi: 10.3390/molecules24030453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang N., Zhang S., Yuan Y., Xu H., Defossa E., Matter H., et al. Molecular basis for inhibiting human glucose transporters by exofacial inhibitors [J] Nat. Commun. 2022;13(1):2632. doi: 10.1038/s41467-022-30326-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mueckler M., Thorens B. The SLC2 (GLUT) family of membrane transporters [J] Mol Aspects Med. 2013;34(2–3):121–138. doi: 10.1016/j.mam.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen L.Q., Cheung L.S., Feng L., Tanner W., Frommer W.B. Transport of sugars [J] Annu. Rev. Biochem. 2015;84:865–894. doi: 10.1146/annurev-biochem-060614-033904. [DOI] [PubMed] [Google Scholar]
- 17.Ohtsubo K., Takamatsu S., Minowa M.T., Yoshida A., Takeuchi M., Marth J.D. Dietary and genetic control of glucose transporter 2 glycosylation promotes insulin secretion in suppressing diabetes [J] Cell. 2005;123(7):1307–1321. doi: 10.1016/j.cell.2005.09.041. [DOI] [PubMed] [Google Scholar]
- 18.Stuart C.A., Howell M.E., Yin D. Overexpression of GLUT5 in diabetic muscle is reversed by pioglitazone [J] Diabetes Care. 2007;30(4):925–931. doi: 10.2337/dc06-1788. [DOI] [PubMed] [Google Scholar]
- 19.Yin Y., Choi S.C., Xu Z., Perry D.J., Seay H., Croker B.P., et al. Normalization of CD4+ T cell metabolism reverses lupus [J] Sci. Transl. Med. 2015;7(274) doi: 10.1126/scitranslmed.aaa0835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jodeleit H., AL-Amodi O., Caesar J., Villarroel Aguilera C., Holdt L., Gropp R., et al. Targeting ulcerative colitis by suppressing glucose uptake with ritonavir [J] Dis Model Mech. 2018;11(11) doi: 10.1242/dmm.036210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tilekar K., Upadhyay N., Iancu C.V., Pokrovsky V., Choe J.Y., Ramaa C.S. Power of two: combination of therapeutic approaches involving glucose transporter (GLUT) inhibitors to combat cancer [J] Biochim. Biophys. Acta Rev. Canc. 2020;1874(2) doi: 10.1016/j.bbcan.2020.188457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morrissey S.M., Zhang F., Ding C., Montoya-Durango D.E., Hu X., Yang C., et al. Tumor-derived exosomes drive immunosuppressive macrophages in a pre-metastatic niche through glycolytic dominant metabolic reprogramming [J] Cell Metab. 2021;33(10):2040. doi: 10.1016/j.cmet.2021.09.002. 58.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Macintyre A.N., Gerriets V.A., Nichols A.G., Michalek R.D., Rudolph M.C., Deoliveira D., et al. The glucose transporter Glut1 is selectively essential for CD4 T cell activation and effector function [J] Cell Metab. 2014;20(1):61–72. doi: 10.1016/j.cmet.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martinez-Outschoorn U.E., Peiris-PagéS M., Pestell R.G., Sotgia F., Lisanti M.P. Cancer metabolism: a therapeutic perspective [J] Nat. Rev. Clin. Oncol. 2017;14(2):113. doi: 10.1038/nrclinonc.2017.1. [DOI] [PubMed] [Google Scholar]
- 25.Bartman C.R., Weilandt D.R., Shen Y., Lee W.D., Han Y., Teslaa T., et al. Slow TCA flux and ATP production in primary solid tumours but not metastases [J] Nature. 2023;614(7947):349–357. doi: 10.1038/s41586-022-05661-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ceballos J., Schwalfenberg M., Karageorgis G., Reckzeh E.S., Sievers S., Ostermann C., et al. Synthesis of indomorphan pseudo-natural product inhibitors of glucose transporters GLUT-1 and -3 [J] Angew Chem. Int. Ed. Engl. 2019;58(47):17016–17025. doi: 10.1002/anie.201909518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goodwin J., Neugent M.L., Lee S.Y., Choe J.H., Choi H., Jenkins D.M.R., et al. The distinct metabolic phenotype of lung squamous cell carcinoma defines selective vulnerability to glycolytic inhibition [J] Nat. Commun. 2017;8 doi: 10.1038/ncomms15503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karageorgis G., Reckzeh E.S., Ceballos J., Schwalfenberg M., Sievers S., Ostermann C., et al. Chromopynones are pseudo natural product glucose uptake inhibitors targeting glucose transporters GLUT-1 and -3 [J] Nat. Chem. 2018;10(11):1103–1111. doi: 10.1038/s41557-018-0132-6. [DOI] [PubMed] [Google Scholar]
- 29.Wu Q., Ba-Alawi W., Deblois G., Cruickshank J., Duan S., Lima-Fernandes E., et al. GLUT1 inhibition blocks growth of RB1-positive triple negative breast cancer [J] Nat. Commun. 2020;11(1):4205. doi: 10.1038/s41467-020-18020-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Flavahan W.A., Wu Q., Hitomi M., Rahim N., Kim Y., Sloan A.E., et al. Brain tumor initiating cells adapt to restricted nutrition through preferential glucose uptake [J] Nat. Neurosci. 2013;16(10):1373–1382. doi: 10.1038/nn.3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Granchi C., Fortunato S., Minutolo F. Anticancer agents interacting with membrane glucose transporters [J] Medchemcomm. 2016;7(9):1716–1729. doi: 10.1039/C6MD00287K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barros L.F., Ruminot I., Sotelo-Hitschfeld T., Lerchundi R., FernáNDEZ-Moncada I. Metabolic recruitment in brain tissue [J] Annu. Rev. Physiol. 2023;85:115–135. doi: 10.1146/annurev-physiol-021422-091035. [DOI] [PubMed] [Google Scholar]
- 33.Bacos K., Perfilyev A., Karagiannopoulos A., Cowan E., Ofori J.K., Bertonnier-Brouty L., et al. Type 2 diabetes candidate genes, including PAX5, cause impaired insulin secretion in human pancreatic islets [J] J. Clin. Invest. 2023;133(4) doi: 10.1172/JCI163612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Low B.S.J., Lim C.S., Ding S.S.L., Tan Y.S., Ng N.H.J., Krishnan V.G., et al. Decreased GLUT2 and glucose uptake contribute to insulin secretion defects in MODY3/HNF1A hiPSC-derived mutant β cells [J] Nat. Commun. 2021;12(1):3133. doi: 10.1038/s41467-021-22843-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shearer A.M., Wang Y., Fletcher E.K., Rana R., Michael E.S., Nguyen N., et al. PAR2 promotes impaired glucose uptake and insulin resistance in NAFLD through GLUT2 and Akt interference [J] Hepatology. 2022;76(6):1778–1793. doi: 10.1002/hep.32589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beckerman M., Harel C., Michael I., Klip A., Bilan P.J., Gallagher E.J., et al. GLUT4-overexpressing engineered muscle constructs as a therapeutic platform to normalize glycemia in diabetic mice [J] Sci. Adv. 2021;7(42) doi: 10.1126/sciadv.abg3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pei J.P., Zhang C.D., Yusupu M., Zhang C., Dai D.Q. Screening and validation of the hypoxia-related signature of evaluating tumor immune microenvironment and predicting prognosis in gastric cancer [J] Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.705511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goldman M.J., Craft B., Hastie M., Repečka K., Mcdade F., Kamath A., et al. Visualizing and interpreting cancer genomics data via the Xena platform [J] Nat. Biotechnol. 2020;38(6):675–678. doi: 10.1038/s41587-020-0546-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baghban R., Roshangar L., Jahanban-Esfahlan R., Seidi K., Ebrahimi-Kalan A., Jaymand M., et al. Tumor microenvironment complexity and therapeutic implications at a glance [J] Cell Commun. Signal. 2020;18(1):59. doi: 10.1186/s12964-020-0530-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thorsson V., Gibbs D.L., Brown S.D., Wolf D., Bortone D.S., Ou Yang T.H., et al. The immune landscape of cancer [J] Immunity. 2018;48(4):812. doi: 10.1016/j.immuni.2018.03.023. 30.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peng Q., Hao L.Y., Guo Y.L., Zhang Z.Q., Ji J.M., Xue Y., et al. Solute carrier family 2 members 1 and 2 as prognostic biomarkers in hepatocellular carcinoma associated with immune infiltration [J] World J Clin Cases. 2022;10(13):3989–4019. doi: 10.12998/wjcc.v10.i13.3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang L., Zhu Z., Liang Q., Tao Y., Jin G., Zhong Y., et al. A novel small molecule glycolysis inhibitor WZ35 exerts anti-cancer effect via metabolic reprogramming [J] J. Transl. Med. 2022;20(1):530. doi: 10.1186/s12967-022-03758-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang Z., Li B., Guo Y., Wu L., Kou F., Yang L. Signatures of multi-omics reveal distinct tumor immune microenvironment contributing to immunotherapy in lung adenocarcinoma [J] Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.723172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Y., Wen H., Sun D. SLC2A1 plays a significant prognostic role in lung adenocarcinoma and is associated with tumor immunity based on bioinformatics analysis [J] Ann. Transl. Med. 2022;10(9):519. doi: 10.21037/atm-22-1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y., Qin H., Bian J., Ma Z., Yi H. SLC2As as diagnostic markers and therapeutic targets in LUAD patients through bioinformatic analysis [J] Front. Pharmacol. 2022;13 doi: 10.3389/fphar.2022.1045179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao R., Ding D., Ding Y., Han R., Wang X., Zhu C. Predicting differences in treatment response and survival time of lung adenocarcinoma patients based on a prognostic risk model of glycolysis-related genes [J] Front. Genet. 2022;13 doi: 10.3389/fgene.2022.828543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li F., He C., Yao H., Liang W., Ye X., Ruan J., et al. GLUT1 regulates the tumor immune microenvironment and promotes tumor metastasis in pancreatic adenocarcinoma via ncRNA-mediated network [J] J. Cancer. 2022;13(8):2540–2558. doi: 10.7150/jca.72161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wei W., Cao B., Xu D., Liu Y., Zhang X., Wang Y. Development and validation of a prognostic prediction model for iron metabolism-related genes in patients with pancreatic adenocarcinoma [J] Front. Genet. 2022;13 doi: 10.3389/fgene.2022.1058062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smeland E., Kilvaer T.K., Sorbye S., Valkov A., Andersen S., Bremnes R.M., et al. Prognostic impacts of hypoxic markers in soft tissue sarcoma [J] Sarcoma. 2012;2012 doi: 10.1155/2012/541650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Heo H.J., Park Y., Lee J.H., Kim Y., Kim E.K., Kim G.H., et al. Clinical big-data-based design of GLUT2-targeted carbon nanodots for accurate diagnosis of hepatocellular carcinoma [J] Nanoscale. 2022;14(45):17053–17064. doi: 10.1039/d2nr04238j. [DOI] [PubMed] [Google Scholar]
- 51.Kim Y.H., Jeong D.C., Pak K., Han M.E., Kim J.Y., Liangwen L., et al. SLC2A2 (GLUT2) as a novel prognostic factor for hepatocellular carcinoma [J] Oncotarget. 2017;8(40):68381–68392. doi: 10.18632/oncotarget.20266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Song Z.B., Yu Y., Zhang G.P., Li S.Q. Genomic instability of mutation-derived gene prognostic signatures for hepatocellular carcinoma [J] Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.728574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fane M., Weeraratna A.T. How the ageing microenvironment influences tumour progression [J] Nat. Rev. Cancer. 2020;20(2):89–106. doi: 10.1038/s41568-019-0222-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Altorki N.K., Markowitz G.J., Gao D., Port J.L., Saxena A., Stiles B., et al. The lung microenvironment: an important regulator of tumour growth and metastasis [J] Nat. Rev. Cancer. 2019;19(1):9–31. doi: 10.1038/s41568-018-0081-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.SautèS-Fridman C., Petitprez F., Calderaro J., Fridman W.H. Tertiary lymphoid structures in the era of cancer immunotherapy [J] Nat. Rev. Cancer. 2019;19(6):307–325. doi: 10.1038/s41568-019-0144-6. [DOI] [PubMed] [Google Scholar]
- 56.Shin E., Koo J.S. Glucose metabolism and glucose transporters in breast cancer [J] Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.728759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Morais M., Dias F., Nogueira I., LeãO A., GonçALVES N., AraúJO L., et al. Cancer cells' metabolism dynamics in renal cell carcinoma patients' outcome: influence of GLUT-1-related hsa-miR-144 and hsa-miR-186 [J] Cancers. 2021;13(7) doi: 10.3390/cancers13071733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Arcidiacono B., Iiritano S., Nocera A., Possidente K., Nevolo M.T., Ventura V., et al. Insulin resistance and cancer risk: an overview of the pathogenetic mechanisms [J] Exp. Diabetes Res. 2012;2012 doi: 10.1155/2012/789174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chung S.J., Nagaraju G.P., Nagalingam A., Muniraj N., Kuppusamy P., Walker A., et al. ADIPOQ/adiponectin induces cytotoxic autophagy in breast cancer cells through STK11/LKB1-mediated activation of the AMPK-ULK1 axis [J] Autophagy. 2017;13(8):1386–1403. doi: 10.1080/15548627.2017.1332565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fuentes-Mattei E., Velazquez-Torres G., Phan L., Zhang F., Chou P.C., Shin J.H., et al. Effects of obesity on transcriptomic changes and cancer hallmarks in estrogen receptor-positive breast cancer [J] J Natl Cancer Inst. 2014;106(7) doi: 10.1093/jnci/dju158. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data included in article/supplementary material/referenced in article.