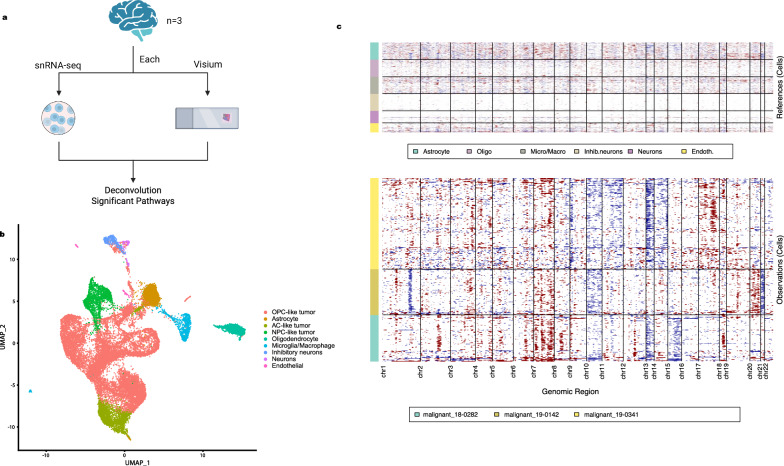
Fig. 1.
a Workflow of the study. From each GBM brain, both snRNA-seq and Visium spatial transcriptomics data were generated as paired samples. The single nuclei data and spatial data were processed separately, then integrated for downstream analysis. b Combined snRNA-seq data from three GBMs with annotation of cell types and tumor cell states. c Combined inferCNV results from three GBMs. Chromosome 7 gain and 10 loss was observed in all samples, and each sample had distinct copy number variations (CNV), demonstrating heterogeneity in CNV