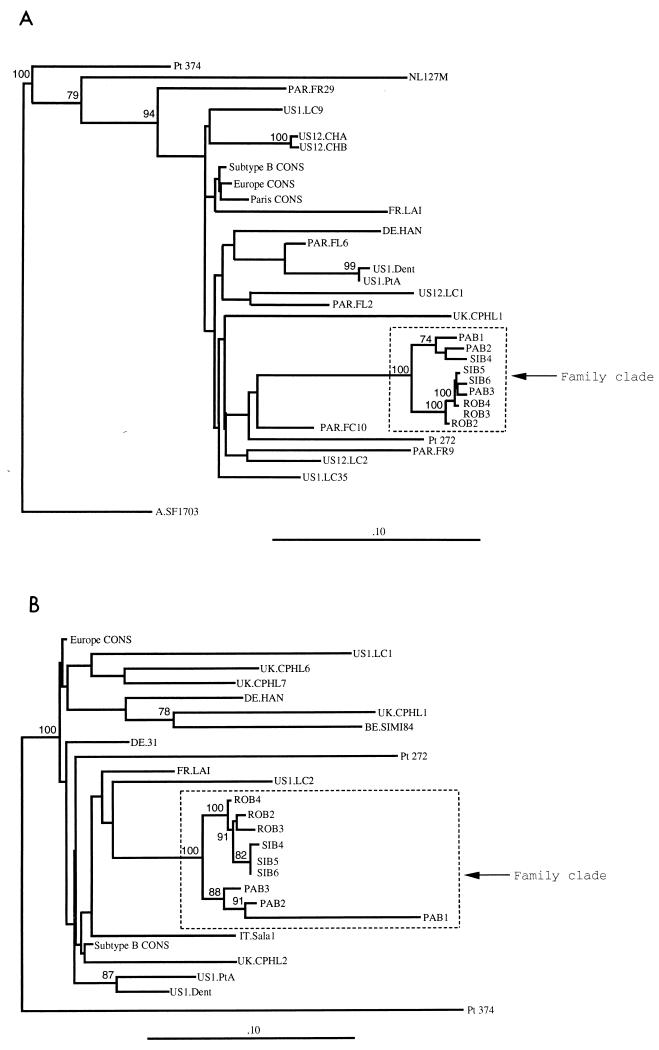
FIG. 3.
Phylogenetic tree analyses comparing the coding sequences for the HIV-1 env C2-V3 region as obtained by the neighbor-joining method (A) and the env V4-C4-V5 region as obtained by the Fitch-Margolias method (B) for the 9 clones derived from proviruses of patients 1 (clones PAB1, PAB2, and PAB3), 3 (clones ROB2, ROB3, and ROB4), and 2 (clones SIB4, SIB5, and SIB6) obtained at a second blood sampling; published sequences of viruses from patients with atypical HIV transmission and their local controls (US) (14, 29); the consensus C2-V3 sequence from 28 controls from Paris (Paris CONS); the consensus C2-V3 and V4-C4-V5 sequences obtained from unrelated controls from Europe (Europe CONS); and the consensus C2-V3 and V4-C4-V5 sequences of HIV-1 subtype B (Subtype B CONS) (32), as previously defined in the legends to Fig. 1 and 2, respectively. An HIV-1 subtype A consensus sequence (A.SF1703), and the V4-C4-V5 sequence of HIV-1 of patient 374 were used as outgroups for the phylogenetic analysis in C2-V3 and V4-C4-V5, respectively.