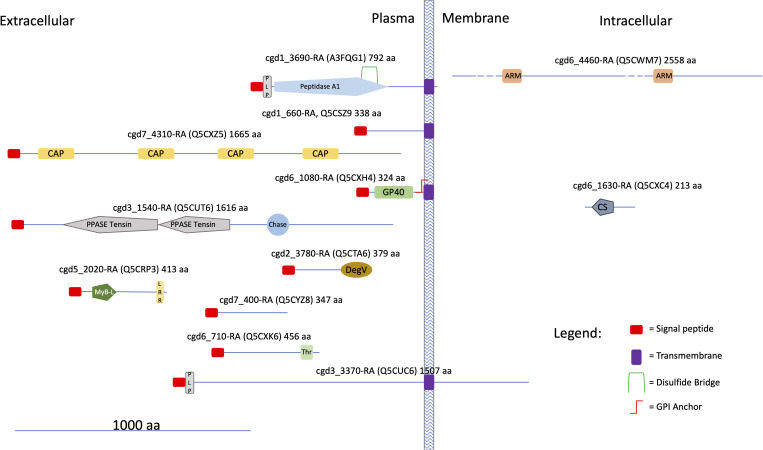
Figure 12.
Cartoon showing some structural features of the Cryptosporidium-characteristic proteins (see text). UniProtKB accessions are reported in brackets. Proteins were divided into three categories according to the presence of hydrophobic motifs (i. e. presence/absence of a signal peptide and/or a transmembrane domain) and their presumptive localization respect to the cell membrane. Prediction of signal peptides and transmembrane domains was performed at Phobius (https://phobius.sbc.su.se/index.html). Prediction of other structural and functional domains was performed at Pfam (http://pfam.xfam.org) and Prosite (https://prosite.expasy.org). List of domains in the figure: Peptidase 1, PEPTIDASE_A1, PS51767; PLP, PROKAR_LIPOPROTEIN, PS51257; CAP, Cysteine-rich secretory protein family, CL0659; GP60, Glycoprotein GP60 of Cryptosporidium, PF11025; PPASE Tensin, PPASE_TENSIN, PS51181; Chase, CHASE, PS50839; DegV, DEGV, PS51482; MyB-L, MYB_LIKE, PS50090; LRR, Leucin Rich Repeat, LRR, PS51450; Thr, Threonin Rich Region, THR_RICH, PS50325; ARM, Armadillo/plakoglobin ARM repeat, ARM_REPEAT, PS50176; CS, CS, PS51203.