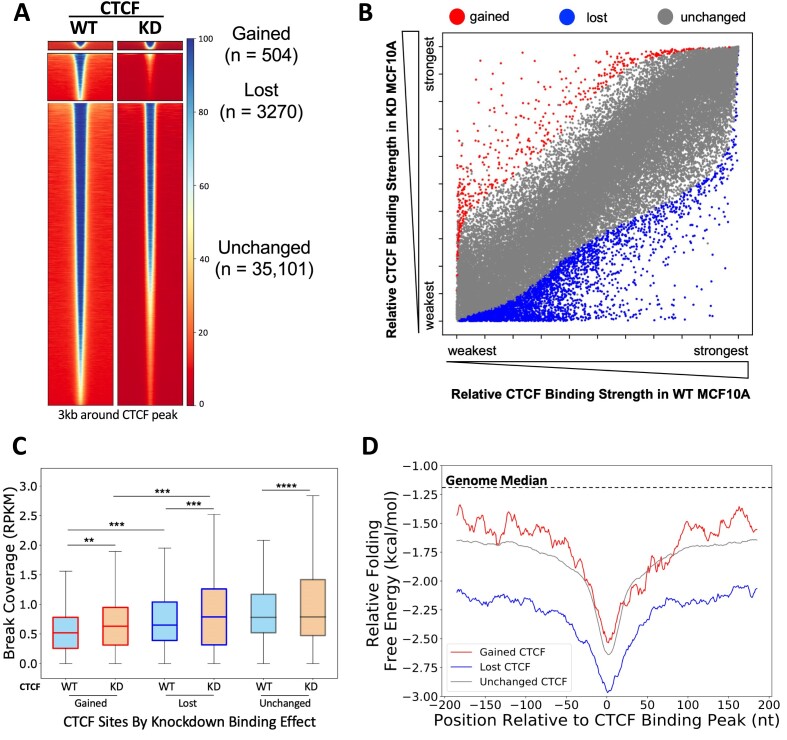
Figure 4.
CTCF knockdown altered binding at CTCF binding sites and differential DSBs and DNA secondary structure folding potentials at gained, lost and unchanged CTCF binding sites in MCF10A cells. (A) Heatmaps show CTCF binding at significantly gained (n = 504), lost (n = 3270) and unchanged (n = 35 101) CTCF binding sites between WT and CTCF knockdown MCF10A cells. Binding peaks with CTCF motifs only were used for the analysis, and differentially binding sites were calculated and identified using DiffBind 3.0 (55). Heatmaps were generated using deepTools (61) at the summit ± 1.5 kb regions of the differential peaks identified by DiffBind. (B) Scatter plot of each CTCF binding site based on relative binding strength rank in WT and CTCF knockdown conditions (gained, red; lost, blue; unchanged, gray). (C) DSBs were mapped in shLuc and shCTCF MCF10A lines. DSBs are significantly increased at gained and lost CTCF binding sites following CTCF knockdown (reads per kilobase million, RPKM). (D) DNA sequences around lost (blue) CTCF binding sites form more energetically favorable alternative DNA secondary structures than unchanged (gray) or gained (red) CTCF binding sites. Boxes denote 25th and 75th percentiles, middle lines show medians and whiskers span from 5% to 95%; **P < 0.01, ***P< 0.001 and ****P ∼ 0; pairwise comparisons, Wilcoxon rank sum test; cross-group comparisons, Kolmogorov–Smirnov test.