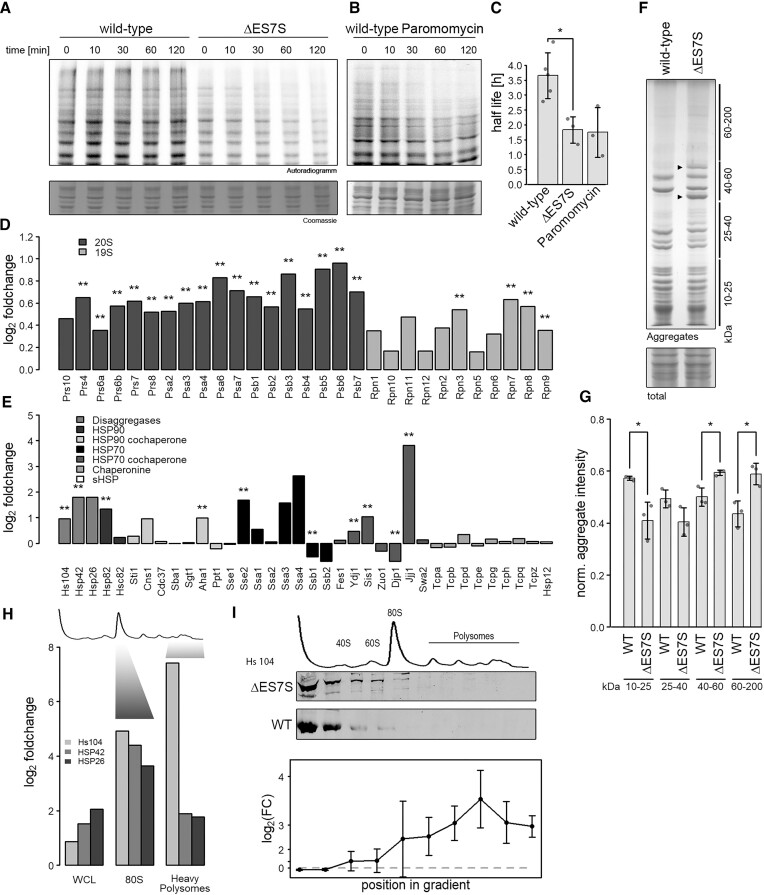
Figure 2.
ΔES7S cells display proteomic instability. Wild-type, ΔES7S cells (A) and wild-type cells pre-treated with 1 mg/ml paromomycin (B) were grown to OD = 0.8, pulsed with radioactive methionine for 10 min and chased with cold methionine for the indicated times. Samples were electrophoresed and radioactivity measured. (C) Decay rates were calculated from the radioactive signal. (D) Proteins constituting the proteasome were investigated via shotgun proteomics. Data are represented as a log2-ratio of ES7S over wild-type cells after quantification with the MaxQuant algorithm (Cox et al., 2014). Stars indicate the significance of foldchanges as determined by Student's t-test and corrected for multiple testing (** P ⇐ 0.01, n = 3). Dark grey bars indicate subunits of the 20S proteasome and light grey those of the 19S cap. (E) Proteins involved in different chaperone networks were investigated via shotgun proteomics. Data are represented as in A (* P ≤ 0.05, ** P ≤ 0.01, n = 3). (F) Cellular aggregates were isolated by repeated rounds of sonication and centrifugation of whole-cell lysates. Aggregates were electrophoresed in 10% polyacrylamide-sodium dodecyl sulfate gels and normalized to whole-cell lysates after staining with colloidal Coomassie. Lines indicate the size range normalized in (F) and arrowheads highlight unique intense bands in ΔES7S aggregates. (G) Intensities of bands in F were quantified and normalized to the intensity of whole-cell lysates. Stars indicate the significance of differences as determined by Student's t-test (n = 3, * P ≤ 0.05). (H) Shotgun proteomics was performed on proteins separated by density-gradient centrifugation. Whole-cell lysates and different fractions corresponding in density to 80S ribosomes and heavy polysomes (as indicated) were analyzed and compared for wild-type and ΔES7S cells. Data are represented as a log2-ratio of ES7S over wild-type cells after quantification with the MaxQuant algorithm. (I) Whole-cell lysates from wild-type and ΔES7S cells were separated by density-gradient centrifugation and fractions were probed for their Hsp104 content via western blotting. Intensities were normalized to uL3 levels and differences were tested for significance by Student t-tests. Data is shown as foldchange ±95% confidence interval.