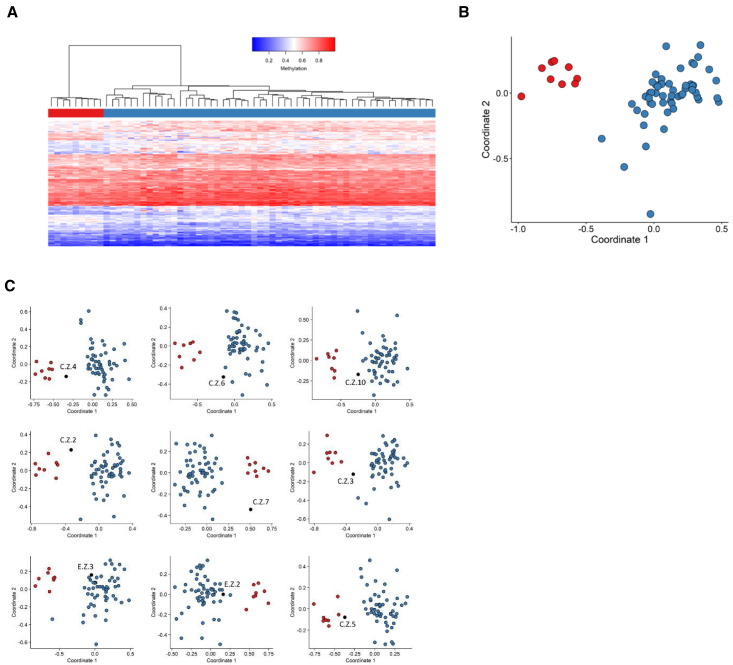
Figure 4.
Assessment of the strength of the identified MKHK-ZZ methylation profile and cross-validation
Using the selected set of probes, unsupervised and supervised models were applied in order to verify the robustness, sensitivity, and specificity of the selected probes in distinguishing ZZ samples from matched control samples.
(A) Hierarchical clustering, where rows represent probes and columns represent samples. The heatmap color scale demonstrates the methylation levels from blue (no methylation) to red (full methylation). On the heatmap pane, red represents ZZ case samples and blue represents control individuals.
(B) MDS, where red and blue circles depict case and control samples, respectively. Plots A and B demonstrate clear separation of the case and control groups.
(C) In order to inspect the sensitivity of the identified ZZ methylation profile, rounds of leave-one-out cross-validation were performed, using all but one ZZ case sample for probe selection at each trial. In each MDS plot, the blue circles represent the matched control samples, red circles indicate ZZ case samples that were used for probe selection, and the black circle depicts the ZZ case sample that was left out from the probe selection process. It was observed that three ZZ samples clustered with control samples when used for testing (C.Z.6, E.Z.3, and E.Z.2) and two samples fell between case and control groups (C.Z.10 and C.Z.3), demonstrating that the selected set of probes are not sensitive enough to classify all ZZ samples correctly. Blue circles represent training samples, while gray circles depict testing samples.